
Epigenetic mechanisms in cancer
History/introduction
A decade and a half ago it would have been unthinkable to see the emergence of headings like “The International Human Epigenome Consortium”, “The NIH Roadmap Epigenomics Mapping Consortium”, “The ENCODE Project”, or the NIH’s National Human Genome Research Institute’s definition of Epigenome: “The epigenome consists of chemical compounds that modify, or mark the genome in a way that tells it what to do, where to do it, and when to do it” (1). After all, it was only 2003 when glorious assessments of the Human Genome Project were flooding scientific and popular publications alike. At the same time, however, the mechanisms for gene regulations were identified as “epigenetic” (2).
The observation that the environment influenced, or sculpted the phenotype of individuals, and these effects were represented in their descendants, goes all the way back, at least to 4th century BC, to Aristoteles (3). In more recent times it was Lamarck (beginning of 19th century), who, in his comprehensive framework for evolution, proposed the idea of inheritance of acquired characteristics, which nowadays is also termed as “soft inheritance” (4). A few years later the genius of Darwin came forward with his proposal on evolution by natural selection. Since those times Lamarckian and Darwinian formulations related to evolution and inheritance, with certain similarities, but also differences, occupied the mainstream debates in biological circles, with the Lamarckian line of thought steadily getting less and less attention (4,5).
The related debates were not only scientific in nature, but were colored with philosophical, religious and political overtones, especially in mid-twentieth century. It was 1942, when C. H. Waddington, a member of the theoretical biology club in Cambridge, UK (6) published what is considered by many the first modern description of epigenetics (7). Ten years later, around 1953, the polemics between the modern synthesis followers, who excluded/opposed the soft inheritance idea, and those who supported it (5) reached a point where ideological allegiances obscured the scientific differences; after all it was the height of the Cold War.
Whereas during the last half of the 20th century the celebrated discovery of the DNA’s double helix structure catalyzed more attention to be paid to the genes and genomics (in 1990 the effort for the Human Genome Project started), in the beginning of this century more attention was given to a multitude of non-genetic influences in biology and society in general. As, unfortunately, it is usual practice in science sometimes to forget, or keep in oblivion important scientific discoveries that set the stage for specific ones to be the celebrated ones (8), in other cases there is a slow but persistent comeback, even from the grave (9).
The vast literature on epigenetics (there were more than 17,000 papers with the root “epigene” between 2010 and 2013) (10) is ever expanding, not only in basic biological systems (11) but in societal aspects, including psychology as well (12). Even dedicated conferences to the subject seem to deal with multiple aspects, from clinical epigenetics (epidemiology, autism) to mechanisms (RNAs as vectors of information transmission) (13).
Generally speaking, epigenetics comprises the studies of variance of gene expression during development and somatic cell proliferation. Or, in other terms, epigenetic mechanisms allow an organism to respond to the environment through changes in gene expression (2). The three most published molecular mechanisms that mediate epigenetic phenomena are DNA methylation, post translational histone modifications, and regulation of non-coding RNAs such as microRNA (miRNA) (14). With respect to cancer, epigenetic approaches to both understand its development, and treatment (14,15).
The increase of knowledge on epigenetics resulted in its reflection on the clinic with new diagnostic and therapeutic strategies. Epigenetics not only offers new insights into the changes in gene regulation that occur during the disease process, but also provides the basis for epigenomics-based targeted therapies. There are many agents currently being tested in clinical trials and some of them are already in clinical use. In addition, drug research and development studies carried out to correct epigenetic errors have gained great speed in recent years. Targeting histone deacetylases (HDACs) and DNA methyltransferase (DNMT)/histone methyltransferase (HMT) are used as possible targets in the treatment of various types of cancer. There are FDA-approved HDAC inhibitors and DNA methylation inhibitors (DNMTIs). Their clinical use gives successful results. Apart from these, histone methylation and miRNAs have also attracted attention as potential therapeutic targets. Combined treatment options of standard chemotherapeutic drugs with epigenetic targeting drugs make it possible to reactivate genes sensitive to chemotherapeutic drugs. It is thought that epigenetic studies will continue for many years and will provide indispensable advantages in many diseases.
This review aims to give a brief summary of the most common epigenetic mechanisms, their possible relations with cancer initiation and progression, focusing on the possible physico-chemical factors that might control these epigenetic mechanisms, and giving examples of the epigenetic therapy approaches. Research articles, books, and other published texts were examined using integrative methodology.
Epigenetic mechanisms of gene regulation
In cancer development, epigenetic mechanisms may directly alter the expression of oncogenes, tumor suppressor genes and other tumor-related genes. Hypermethylation or hypomethylation, especially in the gene promoter regions, global genomic hypomethylation, improper expression of DNMT, histone modification disorders and abnormal expression of non-coding RNAs are the most common epigenetic changes observed in cancer.
DNA and RNA methylation
Among all epigenetic mechanisms, DNA methylation is the most studied. DNMT enzyme catalyze DNA methylation reaction using S-adenosyl methionine as the methyl donor, resulting in 5-methylcytosine (16-18). The most important DNMT enzyme in cancer development is DNMT1 (19). Methylation usually occurs in CpG (cytosine nucleotide followed by a guanine nucleotide) islets clustered in concentrated DNA. While the promoter region that is located at the 5’-end of the human gene is in unmethylated state, the gene is active and allows expression (20). Methylation of this promoter region is usually results in “gene silencing”. Inactivation continues in the daughter cells (21,22). This epigenetic change appears as an alternative pathway to mutation or deletion, which are other causes of gene suppression. Aberrant promoter methylation has been reported to affect several genes regulating cell cycle, adhesion, apoptosis, signal transduction, DNA repair, adhesion and cell differentiation (23,24).
A proportion of 60–80% of the ~29 million CpGs in the human genome are methylated. At least 98% of DNA methylation observed in somatic cells is at a CpG dinucleotide site, while in embryonic stem cells (ESCs) up to a quarter of methylations occur at a non-CpG site. Defects in DNA methylation have been shown to be related with cancer, but no DNMT mutation or deficiency has been identified as a cause of tumor development. Distinctive features of epigenetic changes seen in cancer include global DNA hypomethylation and locus-specific hypermethylation of CpG islands (CGIs) (25). To date, all tumor samples examined show reductions in global DNA methylation (26).
In summary, DNA hypomethylation is mainly observed in heavily methylated repeating body elements and intergenic regions, which causes instability in the genome and activation of oncogenes. Locus-specific hypermethylation, on the other hand, is usually observed in the promoter CGI islets of tumor suppressor genes and results in inherited transcriptional silencing (27).
Unlike DNA methylation, which is a transcriptional modification, RNA methylation is a post-transcriptional modulation. Although it was discovered about 50 years ago, RNA methylation has only recently attracted attention, due to lack of analytical techniques for determining, characterization and sequencing it. There are more than 170 kinds of RNA modifications identified, yet more is expected to follow (28,29). The demethylases, or “erasers” (like ALKBH5, FTO), and methyltransferases, or “writers” (like METTL3/14, KIAA1429 WTAP), and decoder proteins, or “readers” (like YTH, HNRNP) have been discovered more recently, and have led to important discoveries in “epitranscriptomics”. Among other RNA methylations like m1A, m5C or m6Am, the most commonly observed and studied RNA methylation is the N6-methyladenosine (m6A). This notation describes methylation of the adenosine residue at the N-6 position (28-30). These reversible and very dynamic modifications affect all types of coding and non-coding RNAs, therefore are effective in all fundamental cellular processes (30). So, it is no surprise that they have also been found to be associated to several types of cancers, including leukemia, breast, bladder, colorectal, endometrial, hepatocellular, gastrointestinal, lung, liver and pancreas cancers, and epithelial mesenchymal transition (EMT) of metastatic cancer cells (30-32). METTL3, in particular, is now accepted as a biomarker for cancer (30). RNA methylation is shown to also interact with other types of epigenetic modulations, pointing out its strong regulatory role (28). With its broad range of biological effects, RNA methylation modifications have a great potential for unraveling complex cellular mechanisms in health and disease, and are powerful epigenetic therapy targets.
Histone modifications
Histones, the proteins by which the DNA is packaged in chromatin, undergo various post-translational modifications like acetylation, phosphorylation, ubiquitination and ADP ribosylation, usually at their N-terminal tails (33,34). Histone modifications can interact with each other, as well as with DNA methylation, and these interactions can modify the higher order chromatin structure.
Lysine residue acetylation forms chromatin, while their deacetylation correlates to transcriptional repression (35). On the other hand, lysine and arginine residue methylation takes place at histones H3 and H4 by HMTs (36). Different transcriptional events take place based on the location and type of methylated histone. Trithorax group (TrxG) and polycomb group (PcG) of proteins are the two main types of complexes found in histone modifications. Some of these protein groups exhibit HMT activity (37). It has been shown that covalent modifications of histones constitute the link between DNA methylation and gene chromatin silencing (38,39).
miRNAs
MiRNAs are small molecules consisting of approximately 25 nucleotides, synthesized in the nucleus and released into the cytoplasm, binding to mRNAs and causing their expression to change (40). As each miRNA can affect many mRNAs, each mRNA is also under the influence of many miRNAs (41). MiRNAs are encoded within the intergenic regions, introns or exons of protein-coding genes, and they are believed to be co-regulated by host genes (42-44). First a few kb-long primary miRNAs (pri-miRNAs) are synthesized by RNA polymerase II (45,46). Pri-miRNAs are then processed in the nucleus into a precursor miRNA (pre-miRNA) of approximately 70 kb in length (47). The pre-miRNA binds with exportin 5, and is transported from the nucleus to the cytosol. This reaction is catalyzed by a Ran-GTP. Mature miRNA duplex of 22–25 nucleotides in length is synthesized by RNase III enzymes in the cytosol (48,49). After the double-stranded miRNA complex binds to the RNA-induced silencing complex (RISC), the passenger strand is removed while the guide strand remains in the complex and acts as a template to form a new RISC (50). MiRNA molecules can bind to their target genes through the 3' untranslated region (3'UTR), 5'UTR or other gene regions to induce translational repression (51). The “seed” of the miRNA molecule, which is 2–8 nucleotides, binds to the target site, and is essential for functionality and target specificity. The degree of complementarity between the seed and the target is essential for the regulatory mechanism (52,53).
MiRNAs are highly conserved across different species and they regulate several cellular processes, including development, proliferation, differentiation and apoptosis (54-56).
MiRNAs are shown to be linked to cancer, and they can function as oncogenes, as well as tumor suppressor genes (52,57-60). MiRNA expression is epigenetically altered in a tissue-specific manner in both physiological and pathological conditions. They affect the protein levels of target mRNAs without altering their gene sequences. They function at different levels of the genome, can regulate and/or be regulated by other epigenetic actors, such as DNA methylation and histone modifications (61-63), or key enzymes responsible for epigenetic reactions, such as DNMTs, HDACs, and HMTs (64,65). Aberrant methylation patterns of CGIs near or within miRNA genes have been reported to cause a failure in the expression of key miRNAs and resulting pathogenic changes, including tumorigenesis (66). Moreover, some miRNAs can directly affect gene expression at the transcriptional level in the nucleus by complementing the promoter regions of specific genes. In contrast, other miRNAs can affect other chromatin modifiers that cause transcriptional silencing. The interaction between miRNAs and other epigenetic is orchestrated to maintain normal physiological functions. The disruption of this interaction has been associated with many diseases, including cancer (61). CGI hypermethylation that is shown to downregulate tumor suppressor miRNAs is emerging as a common feature of cancer (67,68). On the other hand, hypomethylation of CGIs activates gene expression and has been reported to promote cancer formation (4).
The epigenetic regulation of some miRNAs in cancer types is shown in Table 1.
Table 1
| Cancer types | Target | miRNAs | Function | References |
|---|---|---|---|---|
| Lung | – | miR-1973, miR-494, miR-4286, miR-29b-3p | Reduced apoptosis, higher resistance of chemotherapy | (69) |
| Breast | KDM5B | miR-137 | Migration | (70) |
| Colorectal | TCF4, SUZ12 | miR-145, miR-132, miR-212 | – | (71) |
| Glioma | – | miR-424 | Tumor suppressor, migration, down-regulated by DNA methylation | (72) |
| Gastric | – | miR-196b-5p | Migration, invasion | (73) |
| Lung | CCNE1 | miR‐1179 | Tumor cell growth suppressor | (74) |
| Bladder | DNMT3A, PETN | miR-29 | – | (75) |
Physico-chemical factors on epigenetic mechanisms
Somatic mutations have been identified in many tumor suppressor genes, oncogenes, and cancer-related genes. However, studies have shown that mutations are not sufficient to cause such a disease. When biological processes, such as the formation, proliferation and metastasis of cancer cells are examined at the molecular level, it has been observed that both genetic and epigenetic factors play a role in these biological processes. Epigenetic modifications required for mammalian development and cell proliferation are disrupted by environmental causes. Transcriptional changes occur with disruption of epigenetic processes and malignant cellular transformation is observed. It has been reported that physical and chemical factors, such as environmental pollution, ultraviolet radiation (UVR), nutrition, alcohol and smoking, and physical activity directly affect the disease mechanisms. Understanding the epigenetic mechanisms of these physicochemical factors in the development and progression of cancers will enable us to design our lifestyle and diet, thereby reduce cancer risk and monitor response to treatment. The reversibility of epigenetic modifications may enable the development of new therapeutic strategies targeting these modulations for the prevention and/or treatment of physico-chemically induced cancers. Figure 1 summarizes the possible role of key physico-chemical factors, which along with aspects of everyday lifestyle, influence epigenetic mechanisms.
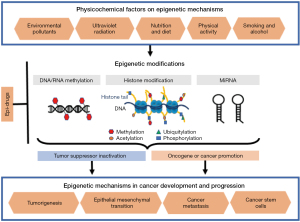
Environmental pollutants
Environmental pollutants, especially air pollutants, heavy metals, organic pollutants and chemicals in drinking water alter gene expression through epigenetic mechanisms (76-79). Air pollution contributes to inflammation and disease development, including cardiovascular diseases, metabolic disorders, asthma and chronic diseases, as well as cancers, due to its harmful components. For example, carbon black (usually from incomplete combustion exhaust gases), nitrogen oxides and polyaromatic hydrocarbons (PAHs) present in polluted air cause decrease in DNA methylation (76,80). PAHs, which are byproducts of the combustion of incomplete fossil fuels or organic elements, are classified as Group 1 carcinogens by the International Agency for Research on Cancer (IARC) (81,82). Most studies of particulate matter and short-term exposure to carbon black have been associated with DNA methylation in steel, oil refinery or petrochemical processing industrial settings (83-86). On the other hand, there are only few studies investigating the effects of other environmental pollutants such as O3, NO2 or SO4 on epigenetic modifications. NO2 has been reported to cause high levels of DNA methylation in the ADRB2 gene (87,88), which is associated with severe asthma patients, while SO4 was associated with decreased LINE-1 methylation (84), and other varying methylation levels in several genes related to asthma (89).
Trace amounts of heavy elements with previously documented carcinogenicity, such as mercury (90,91), arsenic, cadmium (92), and nickel (93), which enter the body in various ways, may lead to genetic and epigenetic changes in different cancer-related genes of somatic and stem cells. For this reason, the underlying epigenetic mechanisms of these trace elements and compounds and their relationship with cancers in heavy metal-contaminated areas are investigated. Examples of studies on epigenetic changes caused by environmental factors are given in Table 2.
Table 2
| Environmental pollutants | Pollutants | Functions/effect | References |
|---|---|---|---|
| Air Pollutant | Particulate matter (PM) | Global DNA hypomethylation, P16 gene promoter hypermethylation, and changes in site specific methylation, acetylation, and phosphorylation of histone H3 | (83,86,87,89,94) |
| Black carbon (BC) | Modulation in allergic asthma gene methylation | (89,95) | |
| Nitrogen dioxide (NO2) | Global DNA methylation or gene specific CpG methylation | (96-99) | |
| Polyaromatic hydrocarbons (PAHs) | Global hypomethylation and hypermethylation of specific genes | (81,82,100) | |
| Heavy metals | Arsenic (As) | Inhibition of DNA methyltransferase and induction of ROS formation |
(91,101) |
| Nickel | Inhibition of DNA hypermethylation (H3K9 mono- and dimethylation), DNMT, and histone H2A, H2B, H3 and H4 acetylation, DNA mutation, ROS generation | (93,102,103) | |
| Mercury (Hg) | Increase of DNA methylation at the promoter region of the glutathione S-transferase mu1 (GSTM1) | (90,104,105) | |
| Cadmium (Cd) | Inhibition of DNMTs activity, DNA hypermethylation and hypomethylation | (92,105-110) | |
| Organic pollutants | Benzene | Decrease of DNA methylation in both Alu and LINE-1 | (111-115) |
| Diethylstilbestrol (DES) | Aberrant DNA methylation of the Hoxa10 gene in utero | (116) | |
| Chemicals in drinking water | Chlorination | Increased carcinogenic risk | (117) |
DNMT, DNA methyltransferase; ROS, reactive oxygen species.
Ultraviolet radiation
Solar UVR is expected to play a significant role in skin tumorigenesis (118,119). Overexposure to solar UVR, especially to ultraviolet B light (UVB, 290–320 nm) component (120), can cause several harmful effects on human skin, including sunburn, photoaging and immunosuppression (121-123). UVB causes DNA damage, epigenetic lesions and irregular gene expression, leading to the main types of skin cancers, including basal cell carcinoma (BCC), squamous cell carcinoma (SCC), and melanoma (124). DNA damage and epigenetic modulations may occur independently, as well as they may affect each other in response to UVR. Studies on biomarkers to distinguish skin lesions have reported hypermethylation of several known tumor suppressor genes related to skin cancers, such as CDH1, CDH3, LAMA3, LAMC2, RASSF1A (125), which causes gene silencing without a change in the coding region. Epigenetic inactivation of RB1/p16 and p53 pathways was also shown in cutaneous SCCs (119). Studies have also shown UVB induced hypoacetylation of histones H3 and H4 in the transcriptionally silenced regions of tumor suppressor genes (126). It is suggested that there is a C→T transition in the di-pyrimidine domains in skin lesions resulting from UVR, and resulting cyclo-butane pyrimidine dimer formation (127). C5-methylation, which occurs at position 5 of cytosine, plays an important role in epigenetic mechanisms involved in the regulation of various biological processes, from cell differentiation to gene expression (128). Recent studies show that ~40% of melanomas are associated with C5-methylation (129,130). As with other environmental factors, DNA methylation is also used as a marker for UVR exposure.
Nutrition and diet
Diet is one of the environmental factors that are more easily studied, and therefore better understood in epigenetic change. There are quite a few dietary components, such as folate, cinnamic acids, polyphenols, resveratrol, cruciferous sulforaphane and isothiocyanates, lignans, selenium and vitamin E are considered to have anti-cancer effects by affecting epigenetic modifications (131-139).
Folate regulates single carbon metabolism that is required for synthesis of DNA, proteins and phospholipids (140). Folate is acquired only through diet. In the body, it is converted to 5,10-methylenetetrahydrofolate (MTHF) which is a significant methyl donor, and is used in methylation of DNA (141-143). Folate deficiency causes a few possible cancer mechanisms, including mutations, DNMT1 inhibition and aberrant global and promoter methylation (135,144-146). Polyphenols that are abundant in plant-derived foods are powerful antioxidants (147), and they inhibit hypermethylation by interacting with the catalytic domain of DNMT1 (116,148). Certain dietary components have similar effects to HDAC inhibitory drugs, causing cell cycle arrest and/or apoptosis in cancer cells (149). A recent study in mice has shown that an omega-3 fatty acid rich maternal diet epigenetically pre-programmed particular genes in the off springs with an increase in acetylation of H3K18 histone and a decrease in H3K4me2 on nucleosomes, that caused a significant protective effect against breast cancer (150).
To investigate the effects of a food on epigenetic changes and disease risk, the intake of that food has to be evaluated in a sufficient number of human samples (78,147). There are studies that support the role of dietary components on epigenetically regulated gene expressions, but the mechanisms of action of these dietary components are still under investigation.
Smoking and alcohol
Cigarette smoke poses a risk for various diseases, such as cardiovascular diseases, chronic obstructive pulmonary disease (COPD) and cancers (151-155). Numerous chemicals in tobacco have toxic effects, including N-nitrosamines, polycyclic aromatic hydrocarbons (benzo[a]pyrene), alkaloids (nicotine and its main metabolite, cotinine), heavy metals (nickel, cadmium, chromium, and arsenic) and aromatic amines (156,157). Smoking causes DNA damage and alteration in DNA methylation and transcription regulation (151,153,157). Altered DNA methylation due to smoking has been studied a lot. Exposure to cigarette smoke raises carbon monoxide levels in the blood, which decreases oxygenation in the body and causes hypoxia, resulting in increased synthesis of S-adenosylmethionine (SAM), one of the main methyl donors, leading to DNA methylation (158).
CYP1A1 and AHRR are among the few genes known to be hypomethylated due to smoking. CYP1A1 is important for the detoxification of carcinogens, while AHRR inhibits the aryl hydrocarbon receptor that metabolizes harmful chemicals. Therefore, inhibition of their functions by hypomethylation increase cancer risk (100,159). Although smoking, in general, leads to decreased DNA methylation, a few critical genes for cell cycle regulation, such as p16 and p53, become hypermethylated due to smoking (160). Loss of function of p16 and p53 can lead to cancer as a result of dysregulation of the cell cycle and uncontrolled cellular divisions (160,161).
Alcohol consumption has been classified as a carcinogenic factor by the IARC and has been associated with various types of cancers (162-166). In general, studies on epigenetic effects of alcohol have focused on DNA methylation and the association of ‘global’ methylation levels with alcohol dependence (166,167). DNA damage induced carcinogenic effect of alcohol is usually associated with ethanol and its metabolite, acetaldehyde, and altered transmethylation reactions (164). Alcohol can also change DNMT activity. While rare alcohol consumption is associated with global hypomethylation, chronic consumption was shown to induce gene-specific DNA hypermethylation, that may lead to cancer. A global increase in histone modifications in oral carcinogenesis has been associated to exposure to ethanol and resulting increases in H3K9/14 and H3K27 acetylation and methylation (168).
Disruption of single carbon metabolic pathway due to alcohol induced by folate deficiency and products of ethanol metabolism has also been reported to cause epigenetic changes associated with cancer development (169,170). Long-term heavy ethanol consumption has been shown to result in elevated homocysteine and S-adenosyl-homocysteine (SAH) levels and decreased SAM and antioxidant glutathione (GSH) levels (143,144,171).
Chronic alcohol consumption has been shown to cause inhibition of the ubiquitin-proteasome pathway in the nucleus, and resulting epigenetic changes. Ethanol metabolism also produces reactive oxygen species (ROS) that can change DNA methylation patterns, increase NADH levels, lead to histone modifications, and induce cancer development (171-174). Chronic high-dose alcohol use also affects some miRNA families, which may also be associated with cancer development (175).
Physical activity
Exercise is strongly emphasized as a strategy for prevention of cancer, as well as for supporting the treatment phase. Yet, epigenetic mechanisms related to exercise are not clearly understood. Physical activity, when done regularly, leads to various epigenetic changes that will benefit cancer patients, such as hypermethylation in the promoter regions for tumor suppressor genes and hypomethylation in the promoter regions of oncogenes (176-178).
The mechanisms that physical activity affects cancer vary depending on age. A large loss of DNA methylation is seen with aging (179) due to the involvement of methyl deoxycytidine, a cytosine methylated at the 5’ carbon of a cytosine (180). The intensity of physical activity is directly related to the amount of promoter demethylation and activation of the expression of many genes (178).
It has been reported that HDACs are highly expressed in muscles, and miRNAs can also be regulated by physical activity (181-183). Furthermore, histone acetylation has been reported to cause selective transcription or inhibition of specific genes related to cancer (184) posttranslational modifications in skeletal muscle (185) or behavioral diseases (180), by specifically modulating H3 and H4. A recent study has shown that sedentary and trained rats with prostate tumors have shown different levels of miR-27a-5p, and exercise increased global DNA methylation while decreasing DNMT expression in the tumor tissue (186). These results support the idea that exercise might reverse the epigenetic modifications due to cancer in tumor tissue.
Stress reduction and lifestyle
Relation between stress and cancer has been well recognized. High stress levels and unhealthy lifestyle reduces the potency of the immune system, which, in turn leaves the people prone to several diseases, including cancer. Involvement of epigenetic factors are also becoming more evident as our understanding of these mechanisms grow. A non-clinical study has shown that a group regularly exercising yoga in order to reduce stress has shown reduced DNA methylation levels in tumor necrosis factor (TNF) regions, along alterations in several other immune system markers (187).
The Lifestyle Medicine Research Summit that was assembled at the University of Pittsburgh on December 4–5, 2019 has determined six core areas of lifestyle medicine that further research needs to focus: plant-predominant nutrition, physical activity, sleep, stress, addictive behaviors, and positive psychology/social connection. All of the determined areas show evidence of epigenetic factors in health and disease and calls for the promise of epigenetic therapy options (188). A recent comprehensive review also gathers literature that correlates epigenetic alterations with lifestyle parameters, such as malnutrition, smoking, high-fat/high-sugar diets, obesity, infections, alcohol consumption, sleep deprivation, chronic stress, air pollution, and chemical exposure, particularly focusing on the effects of epigenetic mechanisms on inflammation (189).
Epigenetic mechanisms in cancer development and progression
Abnormal epigenetic alterations in tumorigenesis
Many types of cancer are associated with aberrant DNA methylation. Global hypomethylation potentially influences cancer tumorigenesis of 5mC-containing sequences, particularly through tumor suppressor genes and deamination of methylated cytosine (190). Coexisting of several epigenetic mechanisms in tumors, including promoter CGI hypermethylation or hypomethylation, and silencing of tumor suppressor genes, suggest that epigenetics might be central to cancer progression (191,192). Promoter hypermethylation has also been reported to up-regulate expression of tumor-promoting genes, such as BCL2 (193), MDR1 (194), HOX11 (195), cMYC (196).
Some DNA methylation changes in cancer are thought to result from mutations in the citric acid cycle. Epigenetic modifications are generally considered as reversible, but some modifications are conserved throughout cancer progression. This provides the advantage that they can be used to classify the disease and predict treatment. In this respect, H3 acetylation and H3K9 di-methylation can identify prostate cancer (PCa) and non-malignant prostate tissue. Similarly, H3K4 tri-methylation is suggested as an important marker of prostate specific antigen (PSA) recurrence (197,198). EZH2 expression, is associated with the aggressiveness of prostate, breast and endometrial cancers (199).
Cancer development includes epigenetic changes both in DNA and chromatin (25,200-202). It is known that promoter DNA hypermethylation of DNA repair genes causes genetic changes (203,204). For example, O(6)-methylguanine-DNA methyltransferase (MGMT), a DNA repair enzyme, can potentially reverse the effects of chemotherapy and radiotherapy. Silencing of MGMT by hypermethylation is suggested to support the treatment (205,206).
Role of epigenetics in EMT and cancer metastasis
The process of epithelial-mesenchymal transition (EMT) in cancer progresses through changes in the morphological features of polarized epithelium, including loss of apical-basal polarity, motility, and cell-cell adhesion (207-209), and acquisition of mesenchymal properties, such as resistance to apoptosis, increased cell motility and invasiveness (208,210,211). Therefore, it is highly associated with metastasis, poor prognosis, blood intravasation (212,213) and resistance to therapy (214). The accumulation of genetic and/or epigenetic changes in tumorigenic cancer precursor cells during cancer development, mostly during the EMT, causes the tumor cells to metastasize to other organs by acquiring a mesenchymal phenotype. EMT is modulated by a variety of stimuli, including tumor-stromal cell interactions, signal (cytokine/growth factor) transduction and hypoxia (215-218).
During EMT, epithelial cells lose E-cadherin expression, resulting in the release of β-catenin (219). Epigenetic mechanisms, such as CpG hypermethylation, acetylation of KLF5 and histone modifications were shown to initiate EMT (220-224). MiRNA-30 was shown to regulate TGF-β and TGF-α induced EMT (225). MiRNAs 143 and 145 were associated with EMT and bone metastasis of PCa (226), while miRNA-1 and miRNA-200 family were shown to inhibit EMT and metastasis by hindering ZEB1 and ZEB2 transcription factors (227-229). p53, a tumor suppressor gene, was shown to regulate EMT through regulating miRNA-200 family (230). Inhibition of miRNA-200 family was shown to increase EMT and metastasis in high-grade breast cancers by increasing H3K27me3-mediated chromatin remodeling and DNA methylation in immortalized human bronchial epithelial cells (227).
DNA methylation of the E-cadherin promoter induces HDACs to be recruited to the region, resulting in histone deacetylation and transcriptional silencing (231). Silencing of EZH2, which is also associated with E-cadherin repression, was also shown to inhibit migratory and invasive characteristics of different cancer cells (232,233), while treatment of some cancer cells with DNMT inhibitors increased their invasiveness, tumorigenicity, and metastatic properties through upregulation of EMT-related genes (234). It has also been reported that histone methylation of the CDH1 promoter also increase invasiveness through suppression of E-cadherin expression (68,235,236).
Role of epigenetics on the cancer stem cell (CSC) model
CSCs, or tumor initiating cells, are a small subpopulation of cells within tumors that show stemness properties, like self-renewal and differentiation, and form the different cell types that make up the heterogeneous tumor mass. CSCs are thought to be responsible for cancer initiation and progression, and resistance to conventional treatment modalities (206,237). There is very little known about the epigenetic regulations in CSCs, but they show quite similar characteristics with ESCs. ESCs are known to maintain their self-renewal abilities even without DNMTs (238,239). However, differentiation of ESCs was found to be almost completely inhibited in the absence of DNMTs. Global DNA hypomethylation was shown to silence pluripotency factors and induce differentiation. ESCs have been reported to preserve their epigenetic memories, and their epigenomes are considered highly stable (240,241).
DNA methylation has also been shown to regulate differentiation of somatic stem cells, like myeloid cells (242), and mesenchymal stem cells (MSCs) (240,243-245). DNA methylation patterns of human MSCs are also found to be quite stable in long-term culture. However, DNA methylation levels differ with aging in regions with H3K9me3, H3K27me3 and EZH2 targets (246,247). Thus, DNA methylation has been suggested as a good molecular marker in characterization of MSCs (247). Epigenetic characterization of CSCs is not well documented in the literature; but it is strongly possible that similar epigenetic control of CSCs with ESCs and MSCs should be observed.
Epigenetic mechanisms have been shown to transform normal stem cells into CSCs when they cause abnormal changes in the differentiation capacity of the stem cells (248,249). For example, isolated human breast CSCs appeared to express low levels of let-7 compared to differentiated breast cancer cells (249). Overexpression of let-7 decreased EZH2 levels and stemness properties of PCa cells, resulting in suppressed clonogenicity and sphere-forming capacity, whereas loss of let-7 increased the expression of EZH2 contributing to PCa invasiveness (250). Since epigenetic mechanisms are regulated under the influence of extracellular changes, they create intratumoral heterogeneity that may promote CSC status. Several miRNAs involved in development are associated with PcG complexes and DNA methylation, and they have an active role in maintaining the balance between self-renewal, proliferation and differentiation in CSCs (52,206,250-253). Thus, epigenetic alterations boost CSC stemness and survival, and contribute to tumor initiation and progression.
Recent studies have reported that, among heterogenous cancer cell populations, those that show both CSC and EMT-like characteristics are more resistant to chemotherapy (254-256). Findings suggest that epigenetic modulations that give these cells CSC and EMT characteristics, as well as abnormal changes in their signaling pathways, such as Wnt/β-catenin or Notch that influence their response to therapy (256-259). Thus, these pathways, and their epigenetic regulations are potential candidates for overcoming drug resistance and CSC targeted therapy.
Current epigenetic treatment approaches and epi-drugs
Early diagnosis in cancer, predicting the prognosis and determining the treatment options is a very laborious and difficult process. “Epigenetic therapy” has become an emerging therapeutic approach (260-263). The fact that epigenetic changes in the genome are reversible provides a new hope for cancer treatment (264). Today, researchers are studying the epigenetic changes seen at different stages of cancer in order to develop new diagnostic and therapeutic approaches for various cancer types. There are FDA approved DNMTIs and HDAC inhibitors, and many more agents that are found to alter methylation patterns or modification of histones on DNA are currently being tested for use in clinics.
DNA methylation inhibitors
Promoter DNA methylation can be used to as a diagnostic marker to molecularly classify cancer, to predict cancer progression, as well as a therapeutic target (265-270). DNMTIs suppress tumor growth and induce apoptosis. Thus, they can restore the activity of tumor suppressor genes. The stability of first generation DNMTIs acting as cytosine analogues is inactivated by cytidine deaminase. Second generation DNMTIs are designed against degradation by cytidine deaminase in order to overcome the stabilization and toxicity problems. Currently available DNMTIs work at the enzymatic level, resulting in global DNA hypomethylation. Although this is therapeutically useful, global hypomethylation has some limitations, such as possibility of oncogene activation and/or increased genomic instability. Promotors located on the repetitive elements in oncogenes can be inactivated by DNA hypomethylation (271). The development of novel DNMTIs targeting specific genes or gene groups, as opposed to global hypomethylation, is a promising approach for more controlled and targeted therapy. Another issue is that DNMTIs are usually activated during the S-phase of the cell cycle. This feature affects fast growing cells, therefore works on highly proliferative cancer cells. However, it does not provide enough clinical benefit in the treatment of diseases that do not have rapid cell cycle. In addition, DNA methylation levels were shown to return to pretreatment levels upon withdrawal (263). Therefore, continuous application is required. A selected list of literature on DNMTIs is given in Table 3. In summary, although DNMTIs are clinically successful, novel inhibitors with higher specificity and cell cycle independence need to be developed to overcome their limitations.
Table 3
| DNMT inhibitors | Class | Generation | Function | Cancer types |
|---|---|---|---|---|
| 5-azacitidine | Nucleoside analogs | First | Sensitizes tumor cells to T-cell-mediated cytotoxicity, enhances efficacy of multiple chemotherapy drugs | Myelomonocytic leukemia (272); lung cancer (273); pancreatic adenocarcinoma (274) |
| 5-aza-2-deoxycytidine (decitabine) | Anti-cancer effect, modulates EMT | Ovarian cancer (275); breast cancer (276); pancreatic cancer (277) | ||
| 5-aza-fluoro-2- deoxycytidine | Second | Inhibits cancer cell proliferation | Colon cancer (278) | |
| 4-deoxyuridine (zebularine) | Inhibits DNA methylation by getting incorporated into DNA | Lung cancer (279); colorectal cancer (280) | ||
| RG108 | Non-nucleoside analogs | First | Induces radio sensitivity, inhibits cell proliferation | Esophageal cancer (281); endometrial cancer (282) |
| EGCG | Suppresses tumor growth, inhibiting NF-κB activation and cell proliferation | Cervical cancer (136); colorectal cancer (137); lung cancer (283) | ||
| Psammaplin | Induces cell cycle arrest, inhibits growth and metastasis | Endometrial cancer (284); lung and glioblastoma (285); breast cancer (286) | ||
| Hydralazine | Induces radio sensitivity | Prostate cancer (287); cervical cancer (288,289) |
DNMT, DNA methyltransferase; EMT, epithelial mesenchymal transition; EGCG, epigallocatechin gallate; NF-κB, nuclear factor kappa B.
HDAC inhibitors
Histone modifications are more unstable than DNA methylation due to the imbalance between histone modifying enzymes (290). So, natural balance can be achieved by correcting the enzyme level in the affected cells. In cancer, it is seen that HMT and histone demethylase enzymes are imbalanced, and there is a global decrease in HDACs (291-296).
Currently, studies are underway to find novel molecules that can minimize the problems with HDACIs by selectively inhibiting specific HDACs, such as HDAC6 and HDAC8 (297-299). A selected list of literature on HDAC inhibitors is given in Table 4. Better exploration of novel and target specific HDACIs will result in more potent therapy options.
Table 4
| HDAC inhibitors | Class | Function | Cancer types |
|---|---|---|---|
| Trichostatin A | Hydroxamic acids | Inhibits cancer cell viability and displays anti-tumor activity | Lung, breast cancer and skin cancer (300); breast cancer (301) |
| SAHA | Inhibits tumor cell growth | Breast cancer (302); pancreatic cancer (303) | |
| Belinostat | Inhibits growth and displays anti-tumor activity | Pancreatic cancer (304) | |
| Resminostat | Blocks platelet-induced HCC cell invasion | Hepatocellular carcinoma (305); pancreatic cancer (306); lung cancer (307) | |
| Abexinostat | Induces CSC differentiation | Breast cancer (308) | |
| Ricolinostat | Increases cancer cell apoptosis | Prostate cancer (309) | |
| Givinostat | Shows anti-proliferative and pro-apoptotic efficacy | Glioblastoma (310) | |
| Valproic acid | Short chain fatty acids | Inhibits cancer cell proliferation by modulating multiple signaling pathways. | Breast cancer (311); thyroid cancer (312); bladder cancer (313) |
| Butyric acid | Displays anti-cancer effect | Gastric cancer (314)colorectal cancer (315); bladder cancer and breast cancer (316) | |
| Entinostat | Benzamides | Displays anti-cancer effect targeting SALL4 | Lung cancer (317); ovarian cancer (318,319) |
| Tacedinaline | Increases cell death | Breast cancer (320) | |
| Mocetinostat | Shows anti-tumor effects | Chondrosarcoma (321); lung cancer (322); breast cancer (323) | |
| 4SC-202 | Inhibits survival and proliferation of cancer cells | Medulloblastoma (324); colorectal cancer (325) | |
| Romidepsin | Cyclic tetrapeptides | Enhances anti-tumor effect, regulates PD-L1 | Bladder cancer (326,327); colon cancer (328) |
| Nicotinamide | Sirtuins inhibitors | Enhances DNA repair and reduces UVR’s immunosuppressive effects | Skin cancer (329,330); cervical cancer (331) |
| Sirtinol | Induces apoptotic effect | Breast cancer (332); lung cancer (333) | |
| Cambinol | Induces antiproliferative effect | Bladder cancer (334) | |
| EX-527 | Increases cell death | Ovarian cancer (335) |
CSC, cancer stem cell; HCC, hepatocellular carcinoma; HDAC, histone deacetylase; SAHA, suberoylanilide hydroxamic acid; UVR, ultraviolet radiation.
Combining epigenetic treatment approaches to increase therapy efficacy has been tried since 1990’s (336). The combined use of DNMT and HDAC inhibition has shown clinical benefits (296,337). Epigenetic drugs can also be combined with conventional treatment options to obtain a stronger response and/or overcome resistance to chemotherapy or radiotherapy. It should be noted that the success of the combination of epigenetic modulations and chemotherapeutic drugs depends on the epigenetic profile of the particular patient and particular cancer type.
Conclusions and future perspectives
Epigenetics plays an important role in cancer development and progression. Early detection of certain epigenetic changes may have predictive and prognostic value in certain cancers. Botanical compounds and pharmaceuticals may be used to modify epigenetic changes to prevent and treat cancer. Integrative oncology includes the use of botanicals, mind-body practices, psychological stress reduction techniques and healthy lifestyle, including physical activity, healthy diet and stress reduction, which have profound epigenetic effects through reduction of oxidative stress and inflammation. Integrative oncology interventions focusing on physical activity, diet and stress reduction may have a role in prevention of cancer development and progression.
Combined treatment options of standard chemotherapeutic drugs with epigenetic approaches can make it possible to reactivate genes sensitive to chemotherapy. Coexistence of multiple epigenetic modifications and the development of drug resistance reveals the necessity of combination therapy in cancer treatment. Chemotherapy resistance may also be overcome by reversing the epigenetic changes that lead to it. Epigenetic therapy approaches further allows for the personalization of the treatment considering the patient’s own history, as well as his/her own lifestyle and health history.
In addition to all its advantages, it can be difficult to work with epigenetic modifications. When DNA methylation biomarkers are analyzed, cell heterogeneity found in tissues from clinical specimens may yield variable, dynamic and complex profiling results. These variable results have to be interpreted in the context of real time changes in the tissues which may in turn influence the epigenetic modifications, which are highly susceptible to local factors, such as cell metabolism, oxygenation, free radical formation and inflammation. In addition, normal cell density is low in body fluids like serum, plasma, urine and sputum, which may make it difficult to detect epigenetic biomarkers in rare cancer cells. However, new highly sensitive cell identification and imaging technologies allow for accurate analysis of single cancer cells found in tissues and circulation, as well as different cell populations in the microenvironment. New methodologies also allow for “liquid biopsies” by identifying cell free DNA in the circulation. It is now possible to get multiple blood and tissues samples during the course of cancer treatment to evaluate genetic and epigenetic changes, which may influence the selection of different anti-cancer treatments.
Heterogeneity in differentiation status in cancerous cells and changes in the histological grade of the tumor over the course of the disease may cause ambiguous results in the correlation of clinical status. While the treatment processes of the disease are being followed, uncertain results may be obtained in patient samples Therefore, clinicians may have to obtain serial blood and tissues samples to understand the changes in genetic and epigenetic profile of the disease, and make necessary treatment changes accordingly. For example, due to the working principle of miRNAs, a miRNA has hundreds of targets. This may cause irregular treatment responses and different pathological processes that appear as a complex network.
In order to overcome the possible problems mentioned above, it is necessary to work in tissues and body fluids with high stability, to purify them at the DNA methylation stage, to create large patient groups, and to use biomarker panels approved by different regulatory agencies, such as FDA or European Medicines Agency (EMA). Also, a good epigenetic biomarker should be more cost-effective than some of the existing markers.
Acknowledgments
Funding: This work was supported by STSM grants from COST Action ‘ENIUS” [CA16217] and COST Action “BIONECA” [CA16122] for Yannis Missirlis and Aylin Sendemir.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Noor N. P. Buchholz) for the series “Integrative Medicine Approaches to Common Urological Problems” published in Longhua Chinese Medicine. The article has undergone external peer review.
Peer Review File: Available at https://lcm.amegroups.com/article/view/10.21037/lcm-21-59/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://lcm.amegroups.com/article/view/10.21037/lcm-21-59/coif). The series “Integrative Medicine Approaches to Common Urological Problems” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
-
Laura Elnitski - Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003;33:245-54. [Crossref] [PubMed]
- Henry D. Aristotle on the mechanism of inheritance. J Hist Biol 2006;39:425-55. [Crossref]
- Handel AE, Ramagopalan SV. Is Lamarckian evolution relevant to medicine? BMC Med Genet 2010;11:73. [Crossref] [PubMed]
- Jablonka E. The evolutionary implications of epigenetic inheritance. Interface Focus 2017;7:20160135. [Crossref] [PubMed]
- Peterson EL. The life organic: The theoretical biology club and the roots of epigenetics. University of Pittsburgh Press, 2017.
- Waddington CH. The epigenotype. 1942. Int J Epidemiol 2012;41:10-3. [Crossref] [PubMed]
- Pray LA. Discovery of DNA structure and function: Watson and Crick. Nature Education 2008;1:100.
- Wang Y, Liu H, Sun Z. Lamarck rises from his grave: parental environment-induced epigenetic inheritance in model organisms and humans. Biol Rev Camb Philos Soc 2017;92:2084-111. [Crossref] [PubMed]
- Burggren WW, Crews D. Epigenetics in comparative biology: why we should pay attention. Integr Comp Biol 2014;54:7-20. [Crossref] [PubMed]
- Arnsdorf EJ, Tummala P, Castillo AB, et al. The epigenetic mechanism of mechanically induced osteogenic differentiation. J Biomech 2010;43:2881-6. [Crossref] [PubMed]
- Svalastog AL, Damjanovicova M. Epigenetics, society, and bio-objects. Croat Med J 2015;56:166-8. [Crossref] [PubMed]
- Bohacek J, Engmann O, Germain PL, et al. Transgenerational epigenetic inheritance: from biology to society-Summary Latsis Symposium Aug 28-30, 2017, Zürich, Switzerland. Environ Epigenet 2018;4:dvy012.
- Seyfried TN, Flores RE, Poff AM, et al. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis 2014;35:515-27. [Crossref] [PubMed]
- Fardi M, Solali S, Farshdousti Hagh M. Epigenetic mechanisms as a new approach in cancer treatment: An updated review. Genes Dis 2018;5:304-11. [Crossref] [PubMed]
- Hoffman AM, Cairns P. Epigenetics of kidney cancer and bladder cancer. Epigenomics 2011;3:19-34. [Crossref] [PubMed]
- Yuan Z, Chen S, Gao C, et al. Development of a versatile DNMT and HDAC inhibitor C02S modulating multiple cancer hallmarks for breast cancer therapy. Bioorg Chem 2019;87:200-8. [Crossref] [PubMed]
- Hervouet E, Peixoto P, Delage-Mourroux R, et al. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin Epigenetics 2018;10:17. [Crossref] [PubMed]
- Harb-de la Rosa A, Acker M, Kumar RA, et al. Epigenetics application in the diagnosis and treatment of bladder cancer. Can J Urol 2015;22:7947-51. [PubMed]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell 2007;128:635-8. [Crossref] [PubMed]
- Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: reading the wrong words. Br J Cancer 2008;98:1881-5. [Crossref] [PubMed]
- Su Y, Fang H, Jiang F. Integrating DNA methylation and microRNA biomarkers in sputum for lung cancer detection. Clin Epigenetics 2016;8:109. [Crossref] [PubMed]
- Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A 1998;95:11891-6. [Crossref] [PubMed]
- Leonhardt H, Cardoso MC. DNA methylation, nuclear structure, gene expression and cancer. J Cell Biochem Suppl 2000;78-83. [Crossref] [PubMed]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006;7:21-33. [Crossref] [PubMed]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 2008;9:465-76. [Crossref] [PubMed]
- Wade PA. Methyl CpG-binding proteins and transcriptional repression. Bioessays 2001;23:1131-7. [Crossref] [PubMed]
- Yao L, Yin H, Hong M, et al. RNA methylation in hematological malignancies and its interactions with other epigenetic modifications. Leukemia 2021;35:1243-57. [Crossref] [PubMed]
- Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 2019;20:608-24. [Crossref] [PubMed]
- Zhou Y, Kong Y, Fan W, et al. Principles of RNA methylation and their implications for biology and medicine. Biomed Pharmacother 2020;131:110731. [Crossref] [PubMed]
- Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer 2019;18:103. [Crossref] [PubMed]
- Xie S, Chen W, Chen K, et al. Emerging roles of RNA methylation in gastrointestinal cancers. Cancer Cell Int 2020;20:585. [Crossref] [PubMed]
- Kouzarides T. Chromatin modifications and their function. Cell 2007;128:693-705. [Crossref] [PubMed]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol 2007;14:1008-16. [Crossref] [PubMed]
- Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 2007;26:5310-8. [Crossref] [PubMed]
- Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol 2009;41:87-95. [Crossref] [PubMed]
- Schuettengruber B, Bourbon HM, Di Croce L, et al. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017;171:34-57. [Crossref] [PubMed]
- Wysocka J, Swigut T, Xiao H, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006;442:86-90. [Crossref] [PubMed]
- Li H, Ilin S, Wang W, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006;442:91-5. [Crossref] [PubMed]
- Ying SY, Chang DC, Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol 2008;38:257-68. [Crossref] [PubMed]
- Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev 2009;28:369-78. [Crossref] [PubMed]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J 2007;26:775-83. [Crossref] [PubMed]
- Gao X, Qiao Y, Han D, et al. Enemy or partner: relationship between intronic micrornas and their host genes. IUBMB Life 2012;64:835-40. [Crossref] [PubMed]
- Lutter D, Marr C, Krumsiek J, et al. Intronic microRNAs support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genomics 2010;11:224. [Crossref] [PubMed]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509-24. [Crossref] [PubMed]
- Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004;23:4051-60. [Crossref] [PubMed]
- Han J, Lee Y, Yeom KH, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 2004;18:3016-27. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015;15:321-33. [Crossref] [PubMed]
- Mah SM, Buske C, Humphries RK, et al. miRNA*: a passenger stranded in RNA-induced silencing complex? Crit Rev Eukaryot Gene Expr 2010;20:141-8. [Crossref] [PubMed]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522-31. [Crossref] [PubMed]
- Brümmer A, Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays 2014;36:617-26. [Crossref] [PubMed]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69. [Crossref] [PubMed]
- Viticchiè G, Lena AM, Latina A, et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle 2011;10:1121-31. [Crossref] [PubMed]
- Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004;303:83-6. [Crossref] [PubMed]
- Zhang J, Guo H, Qian G, et al. MiR-145, a new regulator of the DNA fragmentation factor-45 (DFF45)-mediated apoptotic network. Mol Cancer 2010;9:211. [Crossref] [PubMed]
- Gambari R, Brognara E, Spandidos DA, et al. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: New trends in the development of miRNA therapeutic strategies in oncology Int J Oncol 2016;49:5-32. (Review). [Crossref] [PubMed]
- Naidu S, Magee P, Garofalo M. MiRNA-based therapeutic intervention of cancer. J Hematol Oncol 2015;8:68. [Crossref] [PubMed]
- Ors-Kumoglu G, Gulce-Iz S, Biray-Avci C. Therapeutic microRNAs in human cancer. Cytotechnology 2019;71:411-25. [Crossref] [PubMed]
- Saetrom P, Snøve O Jr, Rossi JJ. Epigenetics and microRNAs. Pediatr Res 2007;61:17R-23R. [Crossref] [PubMed]
- Yao Q, Chen Y, Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol 2019;51:11-7. [Crossref] [PubMed]
- Weber B, Stresemann C, Brueckner B, et al. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle 2007;6:1001-5. [Crossref] [PubMed]
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 2014;13:673-91. [Crossref] [PubMed]
- Li Y, He Q, Wen X, et al. EZH2-DNMT1-mediated epigenetic silencing of miR-142-3p promotes metastasis through targeting ZEB2 in nasopharyngeal carcinoma. Cell Death Differ 2019;26:1089-106. [Crossref] [PubMed]
- Sato S, Katsushima K, Shinjo K, et al. Histone Deacetylase Inhibition in Prostate Cancer Triggers miR-320-Mediated Suppression of the Androgen Receptor. Cancer Res 2016;76:4192-204. [Crossref] [PubMed]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415-28. [Crossref] [PubMed]
- Van den Hove DL, Kompotis K, Lardenoije R, et al. Epigenetically regulated microRNAs in Alzheimer's disease. Neurobiol Aging 2014;35:731-45. [Crossref] [PubMed]
- Mazar J, Khaitan D, DeBlasio D, et al. Epigenetic regulation of microRNA genes and the role of miR-34b in cell invasion and motility in human melanoma. PLoS One 2011;6:e24922. [Crossref] [PubMed]
- El-Awady RA, Hersi F, Al-Tunaiji H, et al. Epigenetics and miRNA as predictive markers and targets for lung cancer chemotherapy. Cancer Biol Ther 2015;16:1056-70. [Crossref] [PubMed]
- Denis H, Van Grembergen O, Delatte B, et al. MicroRNAs regulate KDM5 histone demethylases in breast cancer cells. Mol Biosyst 2016;12:404-13. [Crossref] [PubMed]
- Wang W, Xiao X, Chen X, et al. Tumor-suppressive miR-145 co-repressed by TCF4-β-catenin and PRC2 complexes forms double-negative regulation loops with its negative regulators in colorectal cancer. Int J Cancer 2018;142:308-21. [Crossref] [PubMed]
- Jin C, Li M, Ouyang Y, et al. MiR-424 functions as a tumor suppressor in glioma cells and is down-regulated by DNA methylation. J Neurooncol 2017;133:247-55. [Crossref] [PubMed]
- Shao L, Chen Z, Peng D, et al. Methylation of the HOXA10 Promoter Directs miR-196b-5p-Dependent Cell Proliferation and Invasion of Gastric Cancer Cells. Mol Cancer Res 2018;16:696-706. [Crossref] [PubMed]
- Heller G, Altenberger C, Steiner I, et al. DNA methylation of microRNA-coding genes in non-small-cell lung cancer patients. J Pathol 2018;245:387-98. [Crossref] [PubMed]
- Palmbos PL, Wang L, Yang H, et al. ATDC/TRIM29 Drives Invasive Bladder Cancer Formation through miRNA-Mediated and Epigenetic Mechanisms. Cancer Res 2015;75:5155-66. [Crossref] [PubMed]
- Baudouin C, Charveron M, Tarroux R, et al. Environmental pollutants and skin cancer. Cell Biol Toxicol 2002;18:341-8. [Crossref] [PubMed]
- Rui QY, Li X, Zhang HB, et al. Research on the relationship between environmental chemical pollutant exposure and epigenetics. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 2020;38:237-40. [PubMed]
- Baccarelli A, Cassano PA, Litonjua A, et al. Cardiac autonomic dysfunction: effects from particulate air pollution and protection by dietary methyl nutrients and metabolic polymorphisms. Circulation 2008;117:1802-9. [Crossref] [PubMed]
- Brook RD, Franklin B, Cascio W, et al. Air pollution and cardiovascular disease: a statement for healthcare professionals from the Expert Panel on Population and Prevention Science of the American Heart Association. Circulation 2004;109:2655-71. [Crossref] [PubMed]
- Breton CV, Marutani AN. Air Pollution and Epigenetics: recent Findings. Curr Environ Health Rep 2014;1:35-45. [Crossref]
- Duan H, He Z, Ma J, et al. Global and MGMT promoter hypomethylation independently associated with genomic instability of lymphocytes in subjects exposed to high-dose polycyclic aromatic hydrocarbon. Arch Toxicol 2013;87:2013-22. [Crossref] [PubMed]
- Pavanello S, Pesatori AC, Dioni L, et al. Shorter telomere length in peripheral blood lymphocytes of workers exposed to polycyclic aromatic hydrocarbons. Carcinogenesis 2010;31:216-21. [Crossref] [PubMed]
- Kile ML, Fang S, Baccarelli AA, et al. A panel study of occupational exposure to fine particulate matter and changes in DNA methylation over a single workday and years worked in boilermaker welders. Environ Health 2013;12:47. [Crossref] [PubMed]
- Madrigano J, Baccarelli A, Mittleman MA, et al. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect 2011;119:977-82. [Crossref] [PubMed]
- Peluso M, Bollati V, Munnia A, et al. DNA methylation differences in exposed workers and nearby residents of the Ma Ta Phut industrial estate, Rayong, Thailand. Int J Epidemiol 2012;41:1753-60; discussion 1761-3. [Crossref] [PubMed]
- Dioni L, Hoxha M, Nordio F, et al. Effects of short-term exposure to inhalable particulate matter on telomere length, telomerase expression, and telomerase methylation in steel workers. Environ Health Perspect 2011;119:622-7. [Crossref] [PubMed]
- Breton CV, Salam MT, Wang X, et al. Particulate matter, DNA methylation in nitric oxide synthase, and childhood respiratory disease. Environ Health Perspect 2012;120:1320-6. [Crossref] [PubMed]
- Salam MT, Byun HM, Lurmann F, et al. Genetic and epigenetic variations in inducible nitric oxide synthase promoter, particulate pollution, and exhaled nitric oxide levels in children. J Allergy Clin Immunol 2012;129:232-9.e1-7.
- Sofer T, Baccarelli A, Cantone L, et al. Exposure to airborne particulate matter is associated with methylation pattern in the asthma pathway. Epigenomics 2013;5:147-54. [Crossref] [PubMed]
- Bjørklund G, Pivina L, Dadar M, et al. Mercury Exposure, Epigenetic Alterations and Brain Tumorigenesis: A Possible Relationship? Curr Med Chem 2020;27:6596-610. [Crossref] [PubMed]
- Argos M. Arsenic Exposure and Epigenetic Alterations: Recent Findings Based on the Illumina 450K DNA Methylation Array. Curr Environ Health Rep 2015;2:137-44. [Crossref] [PubMed]
- Wang B, Li Y, Shao C, et al. Cadmium and its epigenetic effects. Curr Med Chem 2012;19:2611-20. [Crossref] [PubMed]
- Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem Res Toxicol 2008;21:28-44. [Crossref] [PubMed]
- Heßelbach K, Kim GJ, Flemming S, et al. Disease relevant modifications of the methylome and transcriptome by particulate matter (PM2.5) from biomass combustion. Epigenetics 2017;12:779-92. [Crossref] [PubMed]
- Jung KH, Lovinsky-Desir S, Yan B, et al. Effect of personal exposure to black carbon on changes in allergic asthma gene methylation measured 5 days later in urban children: importance of allergic sensitization. Clin Epigenetics 2017;9:61. [Crossref] [PubMed]
- Bae JM. Necessity of Epigenetic Epidemiology Studies on the Carcinogenesis of Lung Cancer in Never Smokers. J Prev Med Public Health 2018;51:263-4. [Crossref] [PubMed]
- Sahay D, Terry MB, Miller R. Is breast cancer a result of epigenetic responses to traffic-related air pollution? A review of the latest evidence. Epigenomics 2019;11:701-14. [Crossref] [PubMed]
- Chatkin J, Correa L, Santos U. External Environmental Pollution as a Risk Factor for Asthma. Clin Rev Allergy Immunol 2021; [Epub ahead of print]. [PubMed]
- Lee SY, Chang YS, Cho SH. Allergic diseases and air pollution. Asia Pac Allergy 2013;3:145-54. [Crossref] [PubMed]
- Alhamdow A, Lindh C, Hagberg J, et al. DNA methylation of the cancer-related genes F2RL3 and AHRR is associated with occupational exposure to polycyclic aromatic hydrocarbons. Carcinogenesis 2018;39:869-78. [Crossref] [PubMed]
- Cooper KL, Liu KJ, Hudson LG. Enhanced ROS production and redox signaling with combined arsenite and UVA exposure: contribution of NADPH oxidase. Free Radic Biol Med 2009;47:381-8. [Crossref] [PubMed]
- Genchi G, Carocci A, Lauria G, et al. Nickel: Human Health and Environmental Toxicology. Int J Environ Res Public Health 2020;17:679. [Crossref] [PubMed]
- Sun H, Shamy M, Costa M. Nickel and epigenetic gene silencing. Genes (Basel) 2013;4:583-95. [Crossref] [PubMed]
- Khan F, Momtaz S, Abdollahi M. The relationship between mercury exposure and epigenetic alterations regarding human health, risk assessment and diagnostic strategies. J Trace Elem Med Biol 2019;52:37-47. [Crossref] [PubMed]
- Hanna CW, Bloom MS, Robinson WP, et al. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum Reprod 2012;27:1401-10. [Crossref] [PubMed]
- Wang X, Su P. Cadmium exposure in reproduction: Epigenetic effects. Basic Clin Pharmacol Toxicol 2019;125(Supplement 6):abstr 008.
- Vilahur N, Vahter M, Broberg K. The Epigenetic Effects of Prenatal Cadmium Exposure. Curr Environ Health Rep 2015;2:195-203. [Crossref] [PubMed]
- Venza M, Visalli M, Biondo C, et al. Epigenetic effects of cadmium in cancer: focus on melanoma. Curr Genomics 2014;15:420-35. [Crossref] [PubMed]
- Jiang G, Xu L, Song S, et al. Effects of long-term low-dose cadmium exposure on genomic DNA methylation in human embryo lung fibroblast cells. Toxicology 2008;244:49-55. [Crossref] [PubMed]
- Ray PD, Yosim A, Fry RC. Incorporating epigenetic data into the risk assessment process for the toxic metals arsenic, cadmium, chromium, lead, and mercury: strategies and challenges. Front Genet 2014;5:201. [Crossref] [PubMed]
- Philbrook NA, Winn LM. Investigating the effects of in utero benzene exposure on epigenetic modifications in maternal and fetal CD-1 mice. Toxicol Appl Pharmacol 2015;289:12-9. [Crossref] [PubMed]
- Jiménez-Garza O, Guo L, Byun HM, et al. Promoter methylation status in genes related with inflammation, nitrosative stress and xenobiotic metabolism in low-level benzene exposure: Searching for biomarkers of oncogenesis. Food Chem Toxicol 2017;109:669-76. [Crossref] [PubMed]
- Peng C, Ng JC. The Role of Epigenetic Changes in Benzene- Induced Acute Myeloid Leukaemia. J Clin Epigenet. 2016;2:2. [Crossref]
- Spatari G, Allegra A, Carrieri M, et al. Epigenetic Effects of Benzene in Hematologic Neoplasms: The Altered Gene Expression. Cancers (Basel) 2021;13:2392. [Crossref] [PubMed]
- Fenga C, Gangemi S, Costa C. Benzene exposure is associated with epigenetic changes Mol Med Rep 2016;13:3401-5. (Review). [Crossref] [PubMed]
- Bromer JG, Wu J, Zhou Y, et al. Hypermethylation of homeobox A10 by in utero diethylstilbestrol exposure: an epigenetic mechanism for altered developmental programming. Endocrinology 2009;150:3376-82. [Crossref] [PubMed]
- Deryabkina LA, Marchenko BI, Plugotarenko NK, et al. Assessing Efficiency Of Pre-Ammonization Aimed At Reducing Carcinogenic Risks Caused By Trihalomethanes In Drinking Water. Health Risk Analysis 2020;70-77. [Crossref]
- Moldovan HR, Wittlich M, John SM, et al. Exposure to solar UV radiation in outdoor construction workers using personal dosimetry. Environ Res 2020;181:108967. [Crossref] [PubMed]
- You YH, Pfeifer GP. Similarities in sunlight-induced mutational spectra of CpG-methylated transgenes and the p53 gene in skin cancer point to an important role of 5-methylcytosine residues in solar UV mutagenesis. J Mol Biol 2001;305:389-99. [Crossref] [PubMed]
- Zhu X, Li F, Yang B, et al. Effects of ultraviolet B exposure on DNA methylation in patients with systemic lupus erythematosus. Exp Ther Med 2013;5:1219-25. [Crossref] [PubMed]
- Park HM, Moon E, Kim AJ, et al. Extract of Punica granatum inhibits skin photoaging induced by UVB irradiation. Int J Dermatol 2010;49:276-82. [Crossref] [PubMed]
- Prasad R, Katiyar SK. Crosstalk Among UV-Induced Inflammatory Mediators, DNA Damage and Epigenetic Regulators Facilitates Suppression of the Immune System. Photochem Photobiol 2017;93:930-6. [Crossref] [PubMed]
- Hanson KM, Gratton E, Bardeen CJ. Sunscreen enhancement of UV-induced reactive oxygen species in the skin. Free Radic Biol Med 2006;41:1205-12. [Crossref] [PubMed]
- Katiyar SK, Singh T, Prasad R, et al. Epigenetic alterations in ultraviolet radiation-induced skin carcinogenesis: interaction of bioactive dietary components on epigenetic targets. Photochem Photobiol 2012;88:1066-74. [Crossref] [PubMed]
- Sathyanarayana UG, Moore AY, Li L, et al. Sun exposure related methylation in malignant and non-malignant skin lesions. Cancer Lett 2007;245:112-20. [Crossref] [PubMed]
- Cui X, Wakai T, Shirai Y, et al. Chronic oral exposure to inorganic arsenate interferes with methylation status of p16INK4a and RASSF1A and induces lung cancer in A/J mice. Toxicol Sci 2006;91:372-81. [Crossref] [PubMed]
- Martinez-Fernandez L, Banyasz A, Esposito L, et al. UV-induced damage to DNA: effect of cytosine methylation on pyrimidine dimerization. Signal Transduct Target Ther 2017;2:17021. [Crossref] [PubMed]
- Tommasi S, Denissenko MF, Pfeifer GP. Sunlight induces pyrimidine dimers preferentially at 5-methylcytosine bases. Cancer Res 1997;57:4727-30. [PubMed]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet 2000;16:168-74. [Crossref] [PubMed]
- Tu Y, Dammann R, Pfeifer GP. Sequence and time-dependent deamination of cytosine bases in UVB-induced cyclobutane pyrimidine dimers in vivo. J Mol Biol 1998;284:297-311. [Crossref] [PubMed]
- Hardy TM, Tollefsbol TO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics 2011;3:503-18. [Crossref] [PubMed]
- Meeran SM, Ahmed A, Tollefsbol TO. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin Epigenetics 2010;1:101-16. [Crossref] [PubMed]
- Verma M. Cancer control and prevention by nutrition and epigenetic approaches. Antioxid Redox Signal 2012;17:355-64. [Crossref] [PubMed]
- Chen J, Xu X. Diet, epigenetic, and cancer prevention. Adv Genet 2010;71:237-55. [Crossref] [PubMed]
- Mayne ST, Playdon MC, Rock CL. Diet, nutrition, and cancer: past, present and future. Nat Rev Clin Oncol 2016;13:504-15. [Crossref] [PubMed]
- Wang YQ, Lu JL, Liang YR, et al. Suppressive Effects of EGCG on Cervical Cancer. Molecules 2018;23:2334. [Crossref] [PubMed]
- Luo KW, Xia J, Cheng BH, et al. Tea polyphenol EGCG inhibited colorectal-cancer-cell proliferation and migration via downregulation of STAT3. Gastroenterol Rep (Oxf) 2020;9:59-70. [Crossref] [PubMed]
- Yuasa Y, Nagasaki H, Akiyama Y, et al. DNA methylation status is inversely correlated with green tea intake and physical activity in gastric cancer patients. Int J Cancer 2009;124:2677-82. [Crossref] [PubMed]
- Bishop KS, Ferguson LR. The interaction between epigenetics, nutrition and the development of cancer. Nutrients 2015;7:922-47. [Crossref] [PubMed]
- Steegers-Theunissen RP, Twigt J, Pestinger V, et al. The periconceptional period, reproduction and long-term health of offspring: the importance of one-carbon metabolism. Hum Reprod Update 2013;19:640-55. [Crossref] [PubMed]
- Chen J, Gammon MD, Chan W, et al. One-carbon metabolism, MTHFR polymorphisms, and risk of breast cancer. Cancer Res 2005;65:1606-14. [Crossref] [PubMed]
- Zeisel SH. Epigenetic mechanisms for nutrition determinants of later health outcomes. Am J Clin Nutr 2009;89:1488S-93S. [Crossref] [PubMed]
- Lu SC, Mato JM. Role of methionine adenosyltransferase and S-adenosylmethionine in alcohol-associated liver cancer. Alcohol 2005;35:227-34. [Crossref] [PubMed]
- Mason JB, Choi SW. Effects of alcohol on folate metabolism: implications for carcinogenesis. Alcohol 2005;35:235-41. [Crossref] [PubMed]
- Shanmugham JR, Zavras AI, Rosner BA, et al. Alcohol-folate interactions in the risk of oral cancer in women: a prospective cohort study. Cancer Epidemiol Biomarkers Prev 2010;19:2516-24. [Crossref] [PubMed]
- Blasiak J, Chojnacki J, Pawlowska E, et al. Nutrition in Cancer Therapy in the Elderly-An Epigenetic Connection? Nutrients 2020;12:3366. [Crossref] [PubMed]
- Barrera LN, Cassidy A, Johnson IT, et al. Epigenetic and antioxidant effects of dietary isothiocyanates and selenium: potential implications for cancer chemoprevention. Proc Nutr Soc 2012;71:237-45. [Crossref] [PubMed]
- Johnson IT, Belshaw NJ. Environment, diet and CpG island methylation: epigenetic signals in gastrointestinal neoplasia. Food Chem Toxicol 2008;46:1346-59. [Crossref] [PubMed]
- Rajendran P, Delage B, Dashwood WM, et al. Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: competing actions of 14-3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol Cancer 2011;10:68. [Crossref] [PubMed]
- Abbas A, Witte T, Patterson WL 3rd, et al. Epigenetic Reprogramming Mediated by Maternal Diet Rich in Omega-3 Fatty Acids Protects From Breast Cancer Development in F1 Offspring. Front Cell Dev Biol 2021;9:682593. [Crossref] [PubMed]
- Guida F, Sandanger TM, Castagné R, et al. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum Mol Genet 2015;24:2349-59. [Crossref] [PubMed]
- Shenker NS, Ueland PM, Polidoro S, et al. DNA methylation as a long-term biomarker of exposure to tobacco smoke. Epidemiology 2013;24:712-6. [Crossref] [PubMed]
- Elliott HR, Tillin T, McArdle WL, et al. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin Epigenetics 2014;6:4. [Crossref] [PubMed]
- Teo KK, Ounpuu S, Hawken S, et al. Tobacco use and risk of myocardial infarction in 52 countries in the INTERHEART study: a case-control study. Lancet 2006;368:647-58. [Crossref] [PubMed]
- Zaghlool SB, Al-Shafai M, Al Muftah WA, et al. Association of DNA methylation with age, gender, and smoking in an Arab population. Clin Epigenetics 2015;7:6. [Crossref] [PubMed]
- Zong D, Liu X, Li J, et al. The role of cigarette smoke-induced epigenetic alterations in inflammation. Epigenetics Chromatin 2019;12:65. [Crossref] [PubMed]
- Lee J, Taneja V, Vassallo R. Cigarette smoking and inflammation: cellular and molecular mechanisms. J Dent Res 2012;91:142-9. [Crossref] [PubMed]
- Mahmoud AM, Ali MM. Methyl Donor Micronutrients that Modify DNA Methylation and Cancer Outcome. Nutrients 2019;11:608. [Crossref] [PubMed]
- Philibert RA, Beach SR, Brody GH. Demethylation of the aryl hydrocarbon receptor repressor as a biomarker for nascent smokers. Epigenetics 2012;7:1331-8. [Crossref] [PubMed]
- Taghavi N, Biramijamal F, Sotoudeh M, et al. p16INK4a hypermethylation and p53, p16 and MDM2 protein expression in esophageal squamous cell carcinoma. BMC Cancer 2010;10:138. [Crossref] [PubMed]
- Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis 2010;31:37-49. [Crossref] [PubMed]
- Cao Y, Willett WC, Rimm EB, et al. Light to moderate intake of alcohol, drinking patterns, and risk of cancer: results from two prospective US cohort studies. BMJ 2015;351:h4238. [Crossref] [PubMed]
- Mayne ST, Cartmel B, Kirsh V, et al. Alcohol and tobacco use prediagnosis and postdiagnosis, and survival in a cohort of patients with early stage cancers of the oral cavity, pharynx, and larynx. Cancer Epidemiol Biomarkers Prev 2009;18:3368-74. [Crossref] [PubMed]
- Wang TH, Hsia SM, Shih YH, et al. Association of Smoking, Alcohol Use, and Betel Quid Chewing with Epigenetic Aberrations in Cancers. Int J Mol Sci 2017;18:1210. [Crossref] [PubMed]
- Shingler E, Robles LA, Perry R, et al. Tobacco and alcohol cessation or reduction interventions in people with oral dysplasia and head and neck cancer: systematic review protocol. Syst Rev 2017;6:161. [Crossref] [PubMed]
- Hashibe M, Brennan P, Chuang SC, et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev 2009;18:541-50. [Crossref] [PubMed]
- Smith IM, Mydlarz WK, Mithani SK, et al. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer 2007;121:1724-8. [Crossref] [PubMed]
- Urvalek AM, Osei-Sarfo K, Tang XH, et al. Identification of Ethanol and 4-Nitroquinoline-1-Oxide Induced Epigenetic and Oxidative Stress Markers During Oral Cavity Carcinogenesis. Alcohol Clin Exp Res 2015;39:1360-72. [Crossref] [PubMed]
- Varela-Rey M, Woodhoo A, Martinez-Chantar ML, et al. Alcohol, DNA methylation, and cancer. Alcohol Res 2013;35:25-35. [PubMed]
- Curtis BJ, Zahs A, Kovacs EJ. Epigenetic targets for reversing immune defects caused by alcohol exposure. Alcohol Res 2013;35:97-113. [PubMed]
- Lee TD, Sadda MR, Mendler MH, et al. Abnormal hepatic methionine and glutathione metabolism in patients with alcoholic hepatitis. Alcohol Clin Exp Res 2004;28:173-81. [Crossref] [PubMed]
- Choudhury M, Park PH, Jackson D, et al. Evidence for the role of oxidative stress in the acetylation of histone H3 by ethanol in rat hepatocytes. Alcohol 2010;44:531-40. [Crossref] [PubMed]
- Bardag-Gorce F, French BA, Joyce M, et al. Histone acetyltransferase p300 modulates gene expression in an epigenetic manner at high blood alcohol levels. Exp Mol Pathol 2007;82:197-202. [Crossref] [PubMed]
- You M, Liang X, Ajmo JM, et al. Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am J Physiol Gastrointest Liver Physiol 2008;294:G892-8. [Crossref] [PubMed]
- Dumitrescu RG. Alcohol-Induced Epigenetic Changes in Cancer. Methods Mol Biol 2018;1856:157-72. [Crossref] [PubMed]
- Grazioli E, Dimauro I, Mercatelli N, et al. Physical activity in the prevention of human diseases: role of epigenetic modifications. BMC Genomics 2017;18:802. [Crossref] [PubMed]
- Barrón-Cabrera E, Ramos-Lopez O, González-Becerra K, et al. Epigenetic Modifications as Outcomes of Exercise Interventions Related to Specific Metabolic Alterations: A Systematic Review. Lifestyle Genom 2019;12:25-44. [Crossref] [PubMed]
- Ntanasis-Stathopoulos J, Tzanninis JG, Philippou A, et al. Epigenetic regulation on gene expression induced by physical exercise. J Musculoskelet Neuronal Interact 2013;13:133-46. [PubMed]
- Wilson VL, Smith RA, Ma S, et al. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem 1987;262:9948-51. [Crossref] [PubMed]
- Lavratti C, Dorneles G, Pochmann D, et al. Exercise-induced modulation of histone H4 acetylation status and cytokines levels in patients with schizophrenia. Physiol Behav 2017;168:84-90. [Crossref] [PubMed]
- Riedel S, Radzanowski S, Bowen TS, et al. Exercise training improves high-density lipoprotein-mediated transcription of proangiogenic microRNA in endothelial cells. Eur J Prev Cardiol 2015;22:899-903. [Crossref] [PubMed]
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423-33. [Crossref] [PubMed]
- Nielsen S, Åkerström T, Rinnov A, et al. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS One 2014;9:e87308. [Crossref] [PubMed]
- Zimmer P, Baumann FT, Bloch W, et al. Impact of exercise on pro inflammatory cytokine levels and epigenetic modulations of tumor-competitive lymphocytes in Non-Hodgkin-Lymphoma patients-randomized controlled trial. Eur J Haematol 2014;93:527-32. [Crossref] [PubMed]
- Philp A, Rowland T, Perez-Schindler J, et al. Understanding the acetylome: translating targeted proteomics into meaningful physiology. Am J Physiol Cell Physiol 2014;307:C763-73. [Crossref] [PubMed]
- Dufresne S, Guéritat J, Wong CP, et al. Exercise training as a modulator of epigenetic events in prostate tumors. Prostate Cancer Prostatic Dis 2021; [Epub ahead of print]. [Crossref] [PubMed]
- Harkess KN, Ryan J, Delfabbro PH, et al. Preliminary indications of the effect of a brief yoga intervention on markers of inflammation and DNA methylation in chronically stressed women. Transl Psychiatry 2016;6:e965. [Crossref] [PubMed]
- Vodovotz Y, Barnard N, Hu FB, et al. Prioritized Research for the Prevention, Treatment, and Reversal of Chronic Disease: Recommendations From the Lifestyle Medicine Research Summit. Front Med (Lausanne) 2020;7:585744. [Crossref] [PubMed]
- Ramos-Lopez O, Milagro FI, Riezu-Boj JI, et al. Epigenetic signatures underlying inflammation: an interplay of nutrition, physical activity, metabolic diseases, and environmental factors for personalized nutrition. Inflamm Res 2021;70:29-49. [Crossref] [PubMed]
- Kusmartsev V, Drożdż M, Schuster-Böckler B, et al. Cytosine Methylation Affects the Mutability of Neighboring Nucleotides in Germline and Soma. Genetics 2020;214:809-23. [Crossref] [PubMed]
- Cheishvili D, Boureau L, Szyf M. DNA demethylation and invasive cancer: implications for therapeutics. Br J Pharmacol 2015;172:2705-15. [Crossref] [PubMed]
- Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 2007;39:237-42. [Crossref] [PubMed]
- Stone A, Cowley MJ, Valdes-Mora F, et al. BCL-2 hypermethylation is a potential biomarker of sensitivity to antimitotic chemotherapy in endocrine-resistant breast cancer. Mol Cancer Ther 2013;12:1874-85. [Crossref] [PubMed]
- Caldera V, Mellai M, Annovazzi L, et al. MGMT hypermethylation and MDR system in glioblastoma cancer stem cells. Cancer Genomics Proteomics 2012;9:171-8. [PubMed]
- Jia JS, Spicuglia S. Hypermethylation of CpG island in promoter region of MEIS1 gene and its expression in HOX11(+) acute T lymphoblastic leukemia. Zhonghua Yi Xue Za Zhi 2013;93:3835-40. [PubMed]
- Haertle L, Bittrich M, Potabattula R, et al. Focusing PI and IMiD Resistance in Multiple Myeloma: impact of DNA Methylation. Blood 2018;132:404. [Crossref]
- Ellinger J, Kahl P, von der Gathen J, et al. Global levels of histone modifications predict prostate cancer recurrence. Prostate 2010;70:61-9. [Crossref] [PubMed]
- Lee JH, Yang B, Lindahl AJ, et al. Identifying Dysregulated Epigenetic Enzyme Activity in Castrate-Resistant Prostate Cancer Development. ACS Chem Biol 2017;12:2804-14. [Crossref] [PubMed]
- Bachmann IM, Halvorsen OJ, Collett K, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol 2006;24:268-73. [Crossref] [PubMed]
- Cairns BR. Emerging roles for chromatin remodeling in cancer biology. Trends Cell Biol 2001;11:S15-21. [Crossref] [PubMed]
- Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene 2001;20:3139-55. [Crossref] [PubMed]
- Wolffe AP. Chromatin remodeling: why it is important in cancer. Oncogene 2001;20:2988-90. [Crossref] [PubMed]
- Lahtz C, Pfeifer GP. Epigenetic changes of DNA repair genes in cancer. J Mol Cell Biol 2011;3:51-8. [Crossref] [PubMed]
- Fink D, Aebi S, Howell SB. The role of DNA mismatch repair in drug resistance. Clin Cancer Res 1998;4:1-6. [PubMed]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol 2010;28:1069-78. [Crossref] [PubMed]
- Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol 2010;6:39-51. [Crossref] [PubMed]
- Chapman HA. Epithelial-mesenchymal interactions in pulmonary fibrosis. Annu Rev Physiol 2011;73:413-35. [Crossref] [PubMed]
- Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest 2009;119:1429-37. [Crossref] [PubMed]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014;15:178-96. [Crossref] [PubMed]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420-8. [Crossref] [PubMed]
- Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 2010;29:4741-51. [Crossref] [PubMed]
- Nieto MA. Context-specific roles of EMT programmes in cancer cell dissemination. Nat Cell Biol 2017;19:416-8. [Crossref] [PubMed]
- Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol 2019;29:212-26. [Crossref] [PubMed]
- Lamouille S, Subramanyam D, Blelloch R, et al. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol 2013;25:200-7. [Crossref] [PubMed]
- Nieto MA, Huang RY, Jackson RA, et al. EMT: 2016. Cell 2016;166:21-45. [Crossref] [PubMed]
- Song J. EMT or apoptosis: a decision for TGF-beta. Cell Res 2007;17:289-90. [Crossref] [PubMed]
- Daugherty RL, Gottardi CJ. Phospho-regulation of Beta-catenin adhesion and signaling functions. Physiology (Bethesda) 2007;22:303-9. [Crossref] [PubMed]
- Kim T, Veronese A, Pichiorri F, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med 2011;208:875-83. [Crossref] [PubMed]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 2007;7:415-28. [Crossref] [PubMed]
- Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004;117:927-39. [Crossref] [PubMed]
- Lombaerts M, van Wezel T, Philippo K, et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer 2006;94:661-71. [Crossref] [PubMed]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009;9:265-73. [Crossref] [PubMed]
- Zhang B, Li Y, Wu Q, et al. Acetylation of KLF5 maintains EMT and tumorigenicity to cause chemoresistant bone metastasis in prostate cancer. Nat Commun 2021;12:1714. [Crossref] [PubMed]
- Li Y, Zhang B, Xiang L, et al. TGF-β causes Docetaxel resistance in Prostate Cancer via the induction of Bcl-2 by acetylated KLF5 and Protein Stabilization. Theranostics 2020;10:7656-70. [Crossref] [PubMed]
- Zhang J, Zhang H, Liu J, et al. miR-30 inhibits TGF-β1-induced epithelial-to-mesenchymal transition in hepatocyte by targeting Snail1. Biochem Biophys Res Commun 2012;417:1100-5. [Crossref] [PubMed]
- Peng X, Guo W, Liu T, et al. Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS One 2011;6:e20341. [Crossref] [PubMed]
- Tellez CS, Juri DE, Do K, et al. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res 2011;71:3087-97. [Crossref] [PubMed]
- Zhang J, Zhang H, Qin Y, et al. MicroRNA-200c-3p/ZEB2 loop plays a crucial role in the tumor progression of prostate carcinoma. Ann Transl Med 2019;7:141. [Crossref] [PubMed]
- Liu YN, Yin JJ, Abou-Kheir W, et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 2013;32:296-306. [Crossref] [PubMed]
- Chang CJ, Chao CH, Xia W, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 2011;13:317-23. [Crossref] [PubMed]
- Koizume S, Tachibana K, Sekiya T, et al. Heterogeneity in the modification and involvement of chromatin components of the CpG island of the silenced human CDH1 gene in cancer cells. Nucleic Acids Res 2002;30:4770-80. [Crossref] [PubMed]
- Cao Q, Yu J, Dhanasekaran SM, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008;27:7274-84. [Crossref] [PubMed]
- Herranz N, Pasini D, Díaz VM, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 2008;28:4772-81. [Crossref] [PubMed]
- Ateeq B, Unterberger A, Szyf M, et al. Pharmacological inhibition of DNA methylation induces proinvasive and prometastatic genes in vitro and in vivo. Neoplasia 2008;10:266-78. [Crossref] [PubMed]
- Brembeck FH, Rosário M, Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr Opin Genet Dev 2006;16:51-9. [Crossref] [PubMed]
- Dey N, Ghosh-Choudhury N, Kasinath BS, et al. TGFβ-stimulated microRNA-21 utilizes PTEN to orchestrate AKT/mTORC1 signaling for mesangial cell hypertrophy and matrix expansion. PLoS One 2012;7:e42316. [Crossref] [PubMed]
- Singh VK, Saini A, Chandra R. The Implications and Future Perspectives of Nanomedicine for Cancer Stem Cell Targeted Therapies. Front Mol Biosci 2017;4:52. [Crossref] [PubMed]
- Jackson M, Krassowska A, Gilbert N, et al. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol 2004;24:8862-71. [Crossref] [PubMed]
- Tsumura A, Hayakawa T, Kumaki Y, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 2006;11:805-14. [Crossref] [PubMed]
- Shipony Z, Mukamel Z, Cohen NM, et al. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature 2014;513:115-9. [Crossref] [PubMed]
- Patil V, Ward RL, Hesson LB. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics 2014;9:823-8. [Crossref] [PubMed]
- Álvarez-Errico D, Vento-Tormo R, Sieweke M, et al. Epigenetic control of myeloid cell differentiation, identity and function. Nat Rev Immunol 2015;15:7-17. [Crossref] [PubMed]
- de Almeida DC, Ferreira MR, Franzen J, et al. Epigenetic Classification of Human Mesenchymal Stromal Cells. Stem Cell Reports 2016;6:168-75. [Crossref] [PubMed]
- Bentivegna A, Roversi G, Riva G, et al. The Effect of Culture on Human Bone Marrow Mesenchymal Stem Cells: Focus on DNA Methylation Profiles. Stem Cells Int 2016;2016:5656701. [Crossref] [PubMed]
- Wagner W, Frobel J, Goetzke R. Epigenetic quality check - how good are your mesenchymal stromal cells? Epigenomics 2016;8:889-94. [Crossref] [PubMed]
- Schellenberg A, Lin Q, Schüler H, et al. Replicative senescence of mesenchymal stem cells causes DNA-methylation changes which correlate with repressive histone marks. Aging (Albany NY) 2011;3:873-88. [Crossref] [PubMed]
- Mendicino M, Bailey AM, Wonnacott K, et al. MSC-based product characterization for clinical trials: an FDA perspective. Cell Stem Cell 2014;14:141-5. [Crossref] [PubMed]
- Büssing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med 2008;14:400-9. [Crossref] [PubMed]
- Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007;131:1109-23. [Crossref] [PubMed]
- Kong D, Heath E, Chen W, et al. Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PLoS One 2012;7:e33729. [Crossref] [PubMed]
- Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res 2010;70:378-87. [Crossref] [PubMed]
- Volinia S, Galasso M, Sana ME, et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci U S A 2012;109:3024-9. [Crossref] [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. [Crossref] [PubMed]
- Bommer GT, Gerin I, Feng Y, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol 2007;17:1298-307. [Crossref] [PubMed]
- Tarasov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 2007;6:1586-93. [Crossref] [PubMed]
- Najafi M, Mortezaee K, Majidpoor J. Cancer stem cell (CSC) resistance drivers. Life Sci 2019;234:116781. [Crossref] [PubMed]
- Li N, Babaei-Jadidi R, Lorenzi F, et al. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis 2019;8:13. [Crossref] [PubMed]
- Burgess RJ, Agathocleous M, Morrison SJ. Metabolic regulation of stem cell function. J Intern Med 2014;276:12-24. [Crossref] [PubMed]
- Wang Z, Li Y, Kong D, et al. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res 2009;69:2400-7. [Crossref] [PubMed]
- Iqbal W, Alkarim S, AlHejin A, et al. Targeting signal transduction pathways of cancer stem cells for therapeutic opportunities of metastasis. Oncotarget 2016;7:76337-53. [Crossref] [PubMed]
- Essex A, Pineda J, Acharya G, et al. Replication Study: Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Elife 2019;8:45426. [Crossref] [PubMed]
- Strauss J, Figg WD. Using Epigenetic Therapy to Overcome Chemotherapy Resistance. Anticancer Res 2016;36:1-4. [PubMed]
- Baretti M, Azad NS. The role of epigenetic therapies in colorectal cancer. Curr Probl Cancer 2018;42:530-47. [Crossref] [PubMed]
- Abdelfatah E, Kerner Z, Nanda N, et al. Epigenetic therapy in gastrointestinal cancer: the right combination. Therap Adv Gastroenterol 2016;9:560-79. [Crossref] [PubMed]
- Egger G, Liang G, Aparicio A, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004;429:457-63. [Crossref] [PubMed]
- You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 2012;22:9-20. [Crossref] [PubMed]
- Soengas MS, Capodieci P, Polsky D, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature 2001;409:207-11. [Crossref] [PubMed]
- Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010;17:510-22. [Crossref] [PubMed]
- Tanemura A, Terando AM, Sim MS, et al. CpG island methylator phenotype predicts progression of malignant melanoma. Clin Cancer Res 2009;15:1801-7. [Crossref] [PubMed]
- Toyota M, Ohe-Toyota M, Ahuja N, et al. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A 2000;97:710-5. [Crossref] [PubMed]
- Shen L, Kondo Y, Ahmed S, et al. Drug sensitivity prediction by CpG island methylation profile in the NCI-60 cancer cell line panel. Cancer Res 2007;67:11335-43. [Crossref] [PubMed]
- Gang AO, Frøsig TM, Brimnes MK, et al. 5-Azacytidine treatment sensitizes tumor cells to T-cell mediated cytotoxicity and modulates NK cells in patients with myeloid malignancies. Blood Cancer J 2014;4:e197. [Crossref] [PubMed]
- Füller M, Klein M, Schmidt E, et al. 5-azacytidine enhances efficacy of multiple chemotherapy drugs in AML and lung cancer with modulation of CpG methylation. Int J Oncol 2015;46:1192-204. [Crossref] [PubMed]
- Gailhouste L, Liew LC, Hatada I, et al. Epigenetic reprogramming using 5-azacytidine promotes an anti-cancer response in pancreatic adenocarcinoma cells. Cell Death Dis 2018;9:468. [Crossref] [PubMed]
- Meng F, Sun G, Zhong M, et al. Anticancer efficacy of cisplatin and trichostatin A or 5-aza-2'-deoxycytidine on ovarian cancer. Br J Cancer 2013;108:579-86. [Crossref] [PubMed]
- Lu A, Wang W, Wang-Renault SF, et al. 5-Aza-2'-deoxycytidine advances the epithelial-mesenchymal transition of breast cancer cells by demethylating Sipa1 promoter-proximal elements. J Cell Sci 2020;133:jcs236125. [Crossref] [PubMed]
- Kong R, Sun G, Li X, et al. Small Molecule Inhibitor C188-9 Synergistically Enhances the Demethylated Activity of Low-Dose 5-Aza-2'-Deoxycytidine Against Pancreatic Cancer. Front Oncol 2020;10:612. [Crossref] [PubMed]
- Zhao Q, Fan J, Hong W, et al. Inhibition of cancer cell proliferation by 5-fluoro-2'-deoxycytidine, a DNA methylation inhibitor, through activation of DNA damage response pathway. Springerplus 2012;1:65. [Crossref] [PubMed]
- You BR, Park WH. Zebularine inhibits the growth of A549 lung cancer cells via cell cycle arrest and apoptosis. Mol Carcinog 2014;53:847-57. [Crossref] [PubMed]
- Yang PM, Lin YT, Shun CT, et al. Zebularine inhibits tumorigenesis and stemness of colorectal cancer via p53-dependent endoplasmic reticulum stress. Sci Rep 2013;3:3219. [Crossref] [PubMed]
- Ou Y, Zhang Q, Tang Y, et al. DNA methylation enzyme inhibitor RG108 suppresses the radioresistance of esophageal cancer. Oncol Rep 2018;39:993-1002. [Crossref] [PubMed]
- Yang L, Hou J, Cui XH, et al. RG108 induces the apoptosis of endometrial cancer Ishikawa cell lines by inhibiting the expression of DNMT3B and demethylation of HMLH1. Eur Rev Med Pharmacol Sci 2017;21:5056-64. [PubMed]
- Zhang L, Chen W, Tu G, et al. Enhanced Chemotherapeutic Efficacy of PLGA-Encapsulated Epigallocatechin Gallate (EGCG) Against Human Lung Cancer. Int J Nanomedicine 2020;15:4417-29. [PubMed]
- Ahn MY, Jung JH, Na YJ, et al. A natural histone deacetylase inhibitor, Psammaplin A, induces cell cycle arrest and apoptosis in human endometrial cancer cells. Gynecol Oncol 2008;108:27-33. [Crossref] [PubMed]
- Wee CW, Kim JH, Kim HJ, et al. Psammaplin A-Modified Novel Radiosensitizers for Human Lung Cancer and Glioblastoma Cells. Journal of Radiation Protection and Research 2019;44:15-25. [Crossref]
- Byun WS, Kim WK, Han HJ, et al. Targeting Histone Methyltransferase DOT1L by a Novel Psammaplin A Analog Inhibits Growth and Metastasis of Triple-Negative Breast Cancer. Mol Ther Oncolytics 2019;15:140-52. [Crossref] [PubMed]
- Graça I, Sousa EJ, Costa-Pinheiro P, et al. Anti-neoplastic properties of hydralazine in prostate cancer. Oncotarget 2014;5:5950-64. [Crossref] [PubMed]
- Mani E, Medina LA, Isaac-Olivé K, et al. Radiosensitization of cervical cancer cells with epigenetic drugs hydralazine and valproate. Eur J Gynaecol Oncol 2014;35:140-2. [PubMed]
- Candelaria M, de la Cruz-Hernandez E, Taja-Chayeb L, et al. DNA methylation-independent reversion of gemcitabine resistance by hydralazine in cervical cancer cells. PLoS One 2012;7:e29181. [Crossref] [PubMed]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 2010;31:27-36. [Crossref] [PubMed]
- Tu B, Zhang M, Liu T, et al. Nanotechnology-Based Histone Deacetylase Inhibitors for Cancer Therapy. Front Cell Dev Biol 2020;8:400. [Crossref] [PubMed]
- Waltregny D, North B, Van Mellaert F, et al. Screening of histone deacetylases (HDAC) expression in human prostate cancer reveals distinct class I HDAC profiles between epithelial and stromal cells. Eur J Histochem 2004;48:273-90. [PubMed]
- Nakagawa M, Oda Y, Eguchi T, et al. Expression profile of class I histone deacetylases in human cancer tissues. Oncol Rep 2007;18:769-74. [Crossref] [PubMed]
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov 2006;5:37-50. [Crossref] [PubMed]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet 2007;8:829-33. [Crossref] [PubMed]
- Kim HJ, Bae SC. Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. Am J Transl Res 2011;3:166-79. [PubMed]
- Bruserud Ø, Stapnes C, Ersvaer E, et al. Histone deacetylase inhibitors in cancer treatment: a review of the clinical toxicity and the modulation of gene expression in cancer cell. Curr Pharm Biotechnol 2007;8:388-400. [Crossref] [PubMed]
- Balasubramanian S, Ramos J, Luo W, et al. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia 2008;22:1026-34. [Crossref] [PubMed]
- Haggarty SJ, Koeller KM, Wong JC, et al. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci U S A 2003;100:4389-94. [Crossref] [PubMed]
- Chang J, Varghese DS, Gillam MC, et al. Differential response of cancer cells to HDAC inhibitors trichostatin A and depsipeptide. Br J Cancer 2012;106:116-25. [Crossref] [PubMed]
- Chen L, Jin T, Zhu K, et al. PI3K/mTOR dual inhibitor BEZ235 and histone deacetylase inhibitor Trichostatin A synergistically exert anti-tumor activity in breast cancer. Oncotarget 2017;8:11937-49. [Crossref] [PubMed]
- Natarajan U, Venkatesan T, Radhakrishnan V, et al. Differential Mechanisms of Cell Death Induced by HDAC Inhibitor SAHA and MDM2 Inhibitor RG7388 in MCF-7 Cells. Cells 2018;8:8. [Crossref] [PubMed]
- Kumagai T, Wakimoto N, Yin D, et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (Vorinostat, SAHA) profoundly inhibits the growth of human pancreatic cancer cells. Int J Cancer 2007;121:656-65. [Crossref] [PubMed]
- Dovzhanskiy DI, Arnold SM, Hackert T, et al. Experimental in vivo and in vitro treatment with a new histone deacetylase inhibitor belinostat inhibits the growth of pancreatic cancer. BMC Cancer 2012;12:226. [Crossref] [PubMed]
- Streubel G, Schrepfer S, Kallus H, et al. Histone deacetylase inhibitor resminostat in combination with sorafenib counteracts platelet-mediated pro-tumoral effects in hepatocellular carcinoma. Sci Rep 2021;11:9587. [Crossref] [PubMed]
- Ikeda M, Ohno I, Ueno H, et al. Phase I study of resminostat, an HDAC inhibitor, combined with S-1 in patients with pre-treated biliary tract or pancreatic cancer. Invest New Drugs 2019;37:109-17. [Crossref] [PubMed]
- Tambo Y, Hosomi Y, Sakai H, et al. Phase I/II study of docetaxel combined with resminostat, an oral hydroxamic acid HDAC inhibitor, for advanced non-small cell lung cancer in patients previously treated with platinum-based chemotherapy. Invest New Drugs 2017;35:217-26. [Crossref] [PubMed]
- Salvador MA, Wicinski J, Cabaud O, et al. The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin Cancer Res 2013;19:6520-31. [Crossref] [PubMed]
- Corno C, Arrighetti N, Ciusani E, et al. Synergistic Interaction of Histone Deacetylase 6- and MEK-Inhibitors in Castration-Resistant Prostate Cancer Cells. Front Cell Dev Biol 2020;8:610. [Crossref] [PubMed]
- Angeletti F, Fossati G, Pattarozzi A, et al. Inhibition of the Autophagy Pathway Synergistically Potentiates the Cytotoxic Activity of Givinostat (ITF2357) on Human Glioblastoma Cancer Stem Cells. Front Mol Neurosci 2016;9:107. [Crossref] [PubMed]
- Wawruszak A, Halasa M, Okon E, et al. Valproic Acid and Breast Cancer: State of the Art in 2021. Cancers (Basel) 2021;13:3409. [Crossref] [PubMed]
- Fu YT, Zheng HB, Zhou L, et al. Valproic acid, targets papillary thyroid cancer through inhibition of c-Met signalling pathway. Am J Transl Res 2017;9:3138-47. [PubMed]
- Liu S, Liang B, Jia H, et al. Evaluation of cell death pathways initiated by antitumor drugs melatonin and valproic acid in bladder cancer cells. FEBS Open Bio 2017;7:798-810. [Crossref] [PubMed]
- Shi X, Gong L, Liu Y, et al. 4-phenylbutyric acid promotes migration of gastric cancer cells by histone deacetylase inhibition-mediated IL-8 upregulation. Epigenetics 2020;15:632-45. [Crossref] [PubMed]
- Pattayil L, Balakrishnan-Saraswathi HT. In Vitro Evaluation of Apoptotic Induction of Butyric Acid Derivatives in Colorectal Carcinoma Cells. Anticancer Res 2019;39:3795-801. [Crossref] [PubMed]
- Shahbazfar AA, Zare P, Ranjbaran M, et al. A survey on anticancer effects of artemisinin, iron, miconazole, and butyric acid on 5637 (bladder cancer) and 4T1 (Breast cancer) cell lines. J Cancer Res Ther 2014;10:1057-62. [Crossref] [PubMed]
- Yong KJ, Li A, Ou WB, et al. Targeting SALL4 by entinostat in lung cancer. Oncotarget 2016;7:75425-40. [Crossref] [PubMed]
- Smith HJ, McCaw TR, Londono AI, et al. The antitumor effects of entinostat in ovarian cancer require adaptive immunity. Cancer 2018;124:4657-66. [Crossref] [PubMed]
- Gupta VG, Hirst J, Petersen S, et al. Entinostat, a selective HDAC1/2 inhibitor, potentiates the effects of olaparib in homologous recombination proficient ovarian cancer. Gynecol Oncol 2021;162:163-72. [Crossref] [PubMed]
- Chu PY, Lai JC, Hou MF, et al. Combination of Tacedinaline and EHMT2 inhibition increases breast cancer cell death involving BIRC5 repression and GADD45A induction. Cancer Res 2019;79:abstr 4715.
- Sheikh TN, Patwardhan P, Schwartz GK. Preclinical study of a combination of mocetinostat (HDAC inhibitor) and 5-AZA-dC (decitabine) in chondrosarcoma. Clin Cancer Res 2018;24:abstr A30.
- Haigentz M, Nemunaitis J, Johnson M, et al. P2.06-008 Phase 1/2 Study of Mocetinostat and Durvalumab (MEDI4736) in Patients with Advanced Solid Tumors and Non Small Cell Lung Cancer (NSCLC). J Thorac Oncol 2017;12:S1073-4. [Crossref]
- Shan W, Jiang Y, Yu H, et al. HDAC2 overexpression correlates with aggressive clinicopathological features and DNA-damage response pathway of breast cancer. Am J Cancer Res 2017;7:1213-26. [PubMed]
- Messerli SM, Hoffman MM, Gnimpieba EZ, et al. 4SC-202 as a Potential Treatment for the Pediatric Brain Tumor Medulloblastoma. Brain Sci 2017;7:147. [Crossref] [PubMed]
- Zhijun H, Shusheng W, Han M, et al. Pre-clinical characterization of 4SC-202, a novel class I HDAC inhibitor, against colorectal cancer cells. Tumour Biol 2016;37:10257-67. [Crossref] [PubMed]
- Fan J, Stanfield J, Guo Y, et al. Effect of trans-2,3-dimethoxycinnamoyl azide on enhancing antitumor activity of romidepsin on human bladder cancer. Clin Cancer Res 2008;14:1200-7. [Crossref] [PubMed]
- Okubo K, Miyai K, Kato K, et al. Simvastatin-romidepsin combination kills bladder cancer cells synergistically. Transl Oncol 2021;14:101154. [Crossref] [PubMed]
- Shi Y, Fu Y, Zhang X, et al. Romidepsin (FK228) regulates the expression of the immune checkpoint ligand PD-L1 and suppresses cellular immune functions in colon cancer. Cancer Immunol Immunother 2021;70:61-73. [Crossref] [PubMed]
- Fania L, Mazzanti C, Campione E, et al. Role of Nicotinamide in Genomic Stability and Skin Cancer Chemoprevention. Int J Mol Sci 2019;20:5946. [Crossref] [PubMed]
- Malesu R, Martin AJ, Lyons JG, et al. Nicotinamide for skin cancer chemoprevention: effects of nicotinamide on melanoma in vitro and in vivo. Photochem Photobiol Sci 2020;19:171-9. [Crossref] [PubMed]
- Hassan RN, Luo H, Jiang W. Effects of Nicotinamide on Cervical Cancer-Derived Fibroblasts: Evidence for Therapeutic Potential. Cancer Manag Res 2020;12:1089-100. [Crossref] [PubMed]
- Wang J, Kim TH, Ahn MY, et al. Sirtinol, a class III HDAC inhibitor, induces apoptotic and autophagic cell death in MCF-7 human breast cancer cells. Int J Oncol 2012;41:1101-9. [Crossref] [PubMed]
- Fong Y, Lin YC, Wu CY, et al. The antiproliferative and apoptotic effects of sirtinol, a sirtuin inhibitor on human lung cancer cells by modulating Akt/β-catenin-Foxo3a axis. ScientificWorldJournal 2014;2014:937051. [Crossref] [PubMed]
- Botta L, Filippi S, Bizzarri BM, et al. Oxidative nucleophilic substitution selectively produces cambinol derivatives with antiproliferative activity on bladder cancer cell lines. Bioorg Med Chem Lett 2019;29:78-82. [Crossref] [PubMed]
- Khaleel EF, Badi RM, Satti HH, et al. Exendin-4 exhibits a tumour suppressor effect in SKOVR-3 and OVACR-3 ovarian cancer cells lines by the activation of SIRT1 and inhibition of NF-κB. Clin Exp Pharmacol Physiol 2020;47:1092-102. [Crossref] [PubMed]
- Cameron EE, Bachman KE, Myöhänen S, et al. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 1999;21:103-7. [Crossref] [PubMed]
- Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol 2009;27:5459-68. [Crossref] [PubMed]
Cite this article as: Ors Kumoglu G, Sendemir A, Tanyolac MB, Bilir B, Kucuk O, Missirlis YF. Epigenetic mechanisms in cancer. Longhua Chin Med 2022;5:4.

